Group Sparse Optimization
An Integrative OMICs Method to Predict Master Transcription Factors for Cell Fate Conversion
Introduction
Cell fate conversion by overexpressing defined factors is a powerful tool in regenerative medicine. However, identifying key factors for cell fate conversion requires laborious experimental efforts, thus many of such conversions have not been achieved yet. Nevertheless, cell fate conversions found in many published studies were incomplete as the expression of important gene sets could not be manipulated thoroughly. Therefore, the identification of master transcription factors for complete and efficient conversion is crucial to render this technology more clinical applicational. In the past decade, systematic analyses on various single-cell and bulk OMICs data have uncovered numerous gene regulatory mechanisms, and made it possible to predict master gene regulators during cell fate conversion. This study introduces a novel computational method predicting master transcription factors based on group sparse optimization, with which can be applicable to both single-cell and bulk OMICs data. When it is compared with other state-of-the-art prediction methods, it demonstrated superior performance. In short, this method facilitates fast identification of key regulators, increases the rate of successful conversion and reduces costs from experimental trials.
The GSO source code, input, output and related OMICs data can be downloaded by clicking the hyperlinks on the left sidebar.
Methods
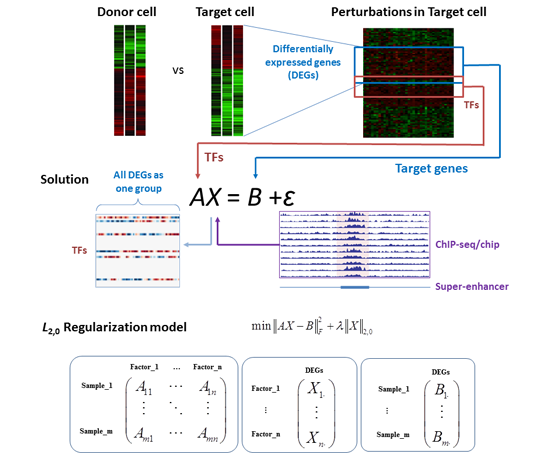
Figure 1. Workflow of the master TF inference. DEGs between donor cells and target cells are identified by comparing expression profiles of two cell types. From transcriptome data of perturbation experiments on target cells, expression profiles of TFs form matrix A, while expression profiles of DEGs form matrix B. The linear model AX = B + ε approximately describes the expression dependency between TFs and DEGs, in which X represents the regulatory strength and ε is the matrix of noise. TF binding and super-enhancer information is transformed into X0 as an initial guess for the solution searching. Master TF inference is an optimization problem to find an X to minimize the difference between AX and B with only a small number of selected TFs, whose regulatory strength on all DEGs are non-zeros. Red color in the solution matrix X means positive regulation, blue color means negative regulation, and white means no regulation. TFs with colors (non-zeros in the solution X) are the predicted master TFs that show regulatory effects on DEGs that need to be changed from donor cells into target cells. The middle panel shows the L2,0 regularization model, while the lower panel lists the structures of matrix A, B and X.
Input
-
A.txtMatrix A: log2-transformed gene expression fold changes between control and TF perturbation samples of 939 regulators in 245 experiments for bulk transcriptome datasets (or 789 regulators in 912 cells for single-cell transcriptome datasets). Candidate regulators, including TFs, mediators, co-factors, chromatin modifiers and repressors, were collected from four TF databases, TRANSFAC, JASPAR, UniPROBE and TFCat, as well as literatures. Each column is the expression profile of each TF in 245 experiments(or 912 cells). Each row is the expression profile of each experiment of all regulators. -
B.txtMatrix B: log2-transformed gene expression fold changes between control and TF perturbation samples of 4000 differentially expressed genes (DEGs) in 245 experiments for bulk transcriptome datasets (or 3636 DEGs in 912 cells for single-cell transcriptome datasets). DEGs are defined by comparing transcriptomes of mouse embryonic fibroblast (MEF) and mouse embryonic stem cells (mESCs). Each column is the expression profile of each target in 245 experiments (or 912 cells). Each row is the expression profile of each experiment of all DEGs. -
InitialX.txtInitial matrixX0describes the connections between TFs and targets, the TF-target connections defined by ChIP-seq/chip data were converted into an initial matrix. Each row is a target gene. Each column is a regulator. If TFihas binding site around the genejpromoter within 10 kbp, the Pearson correlation coefficient (PCC) between the expression profiles of TFiand genejwas calculated and assigned onX0i,j. -
InitialX_SuperEnh.txtInitial matrixX0describes the connections between TFs and targets, the TF-target connections defined by ChIP-seq/chip data were converted into an initial matrix. Each row is a target gene. Each column is a regulator. If TFihas binding site (BS) around the genejpromoter within 10 kbp, the Pearson correlation coefficient (PCC) between the expression profiles of TFiand genejwas calculated and assigned onX0i,j. Super-enhancer regions were used to filter the TFBSs. When a TFBS is outside super-enhancer regions, theX0i,jdefined by this TFBS was reset as 0. -
TFlistL.txt939 candidate regulators in bulk transcriptome datasets (or 789 candidate regulators in single-cell transcriptome datasets), including TFs, mediators, co-factors, chromatin modifiers and repressors, were collected from four TF databases, TRANSFAC, JASPAR, UniPROBE and TFCat, as well as literatures. The order of the 939 (or 789) regulators are the same as those in MatrixAcolumn name, initial matrixXcolumn name and solution matrixXrow name. -
DEGlist.txt4000 DEGs in bulk transcriptome datasets (or 3636 DEGs in single-cell transcriptome datasets). The order of the 4000 (or 3636) DEGs are the same as those in MatrixBrow name, initial matrixXrow name and solution matrixXrow name.
Usage
For single-cell transcriptome data, we have inferred master TFs with four datasets, normalized read counts without imputation and imputed data with three imputation methods, DrImpute, Knn_smooth2 and SAVER. They are saved in four folders Single_cell_Normalized_read_counts, Single_cell_DrImpute, Single_cell_Knn and Single_cell_SAVER. While data of bulk transcriptome is in Bulk_transcriptome. Each folder contains the folder Code, Input and Output.
Infer master TFs using group sparse optimization (GSO) integrating transcriptomes, TF binding and super-enhancer information:
Code files (MaHardThr.m and GSO1.m) are in the folder Code. Input files (A.txt, B.txt and InitialX_SuperEnh.txt) are in the folder Input.
Run GSO1.m via MATLAB.
Then change the directory into folder Output/GSO1. Run the following command to score and rank the predicted TFs:
sh ../../Code/TFScoring.sh Hard
Infer master TFs using GSO integrating transcriptomes and TF binding information:
Code files (MaHardThr.m and GSO2.m) are in the folder Code. Input files (A.txt, B.txt and InitialX.txt) are in the folder Input.
Run GSO2.m via MATLAB.
Then change the directory into folder Output/GSO2. Run the following command to score and rank the predicted TFs:
sh ../../Code/TFScoring.sh Hard
Output
To score and rank the master TFs reported by GSO, we selected a series of group sparsity level K (i.e., the number of TF predicted as master TFs) from 1 to 20, and ran one GSO for each K. For each TF, we used Kmin to denote the smallest value of K when this TF is selected as master TF. We assumed that TFs selected by GSO are more important when K is smaller, so a TF got a higher score if its Kmin was smaller. The score of each TF selected by GSO is defined by

Folders GSO1 and GSO2 are outputs from GSO1.m (GSO integrating transcriptomes, TF binding and super-enhancer information) and GSO2.m (GSO integrating transcriptomes and TF binding information), respectively.
In each output folder, folders s1, s2, s3, s4, s5, s6, s7, s8, s9, s10, s12, s14, s16, s18, s20 contain solution X of each K from 1 to 20. In each sK folder, XHard.txt is the solution matrix X derived from iterative hard thresholding algorithm or iterative soft thresholding algorithm, respectively.Hard_TFranking.txt TF ranking after considering all Ks.
Acknowledgements
This work was supported by National Natural Science Foundation of China (41606143) awarded to JQ, research grants from Research Grants Council, Hong Kong (17121414M), startup funds from Mayo Clinic, USA (Mayo Clinic Arizona and Center for Individualized Medicine) to JW, National Natural Science Foundation of China (11871347), and Natural Science Foundation of Guangdong (2019A1515011917, 2020B1515310008) to YH, and National Science Council of Taiwan (MOST 102-2115-M-039-003-MY3) to JCY.
Contacts
Dr. Jing Qin, School of Pharmaceutical Sciences (Shenzhen), Sun Yat-sen University. Email: qinj29@mail.sysu.edu.cn
Dr. Junwen Wang, Department of Health Sciences Research and Center for Individualized Medicine, Mayo Clinic.
Email: Wang.Junwen@mayo.edu
Dr. Yaohua Hu, College of Mathematics and Statistics, Shenzhen University.
Email: mayhhu@szu.edu.cn