RARG transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1091
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: zeranol
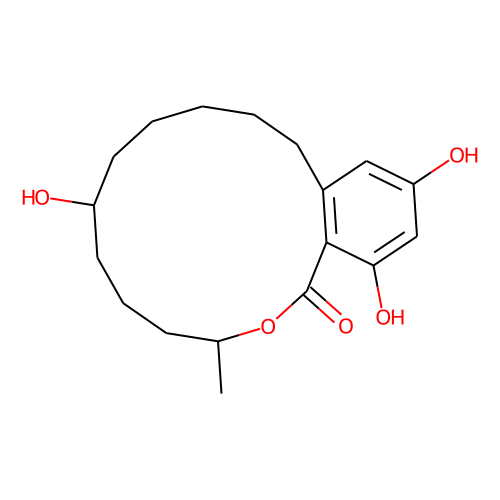
| ID | BRD-A24381660 |
| name | zeranol |
| type | Small molecule |
| molecular formula | C18H26O5 |
| molecular weight | 322.4 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | C[C@H]1CCC[C@@H](CCCCCC2=C(C(=CC(=C2)O)O)C(=O)O1)O |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: RARG
Transcription regulatory factors corresponding to this network
ARTICLE: zeranol
[1] Zeranol: doping offence or mycotoxin? A case-related study.
[2] ERK activation by zeranol has neuroprotective effect in cerebral ischemia reperfusion.
[4] Zeranol--a 'nature-identical' oestrogen?
[7] Formation of Zearalenone Metabolites in Tempeh Fermentation.
[9] Analytical methods for the determination of zeranol residues in animal products: a review.
[10] Identification of a Potent Enzyme for the Detoxification of Zearalenone.
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| CYP2A6 | 7.645 | 8.019 | 8.5263 | 5.6266 | 6.5595 |
| TTYH1 | 8.3338 | 8.57 | 8.4131 | 7.3973 | 7.4432 |
| ZNF16 | 6.6551 | 7.1308 | 6.8138 | 4.8273 | 5.7163 |
| BRD9 | 9.5287 | 9.4086 | 9.2132 | 7.2161 | 8.4273 |
| RIPK4 | 8.5459 | 8.4225 | 8.4304 | 5.1452 | 6.9275 |
| OPLAH | 6.4011 | 6.3741 | 6.0503 | 3.8371 | 5.1793 |
| EIF4G1 | 8.2519 | 8.4944 | 8.4684 | 7.0402 | 7.3693 |
| NUP62 | 8.2342 | 7.8679 | 8.5177 | 5.7004 | 7.2583 |
| RAD9A | 7.2572 | 7.2223 | 7.2361 | 6.1 | 6.1773 |
| SCRN1 | 8.4792 | 8.1337 | 8.1803 | 6.0032 | 7.3273 |
| SMARCA4 | 7.2081 | 7.0522 | 7.4589 | 4.0607 | 6.2207 |
| PRKACA | 10.4363 | 11.0488 | 10.6189 | 6.7881 | 9.2867 |
| PUF60 | 14.2033 | 13.9283 | 13.6941 | 4.6102 | 7.4024 |
| LSM5 | 13.9657 | 13.6053 | 13.6289 | 3.5132 | 5.3375 |
| GRN | 10.1787 | 10.7544 | 11.0535 | 7.9375 | 8.3181 |
| VPS28 | 10.9291 | 10.5969 | 10.1167 | 4.6102 | 4.1423 |
| PTPRK | 11.2223 | 11.3322 | 10.999 | 9.1575 | 7.5823 |
| RAB1B | 9.5829 | 9.4986 | 10.0029 | 5.92 | 8.2372 |
| ASL | 8.3435 | 7.6686 | 8.4944 | 4.436 | 6.8844 |
| CD14 | 7.0683 | 8.3993 | 7.1279 | 4.9496 | 6.103 |
| SART1 | 8.3372 | 8.6723 | 8.4473 | 6.342 | 7.6023 |
| P4HB | 10.8119 | 10.9303 | 10.2065 | 5.9409 | 8.8426 |
| S100A11 | 11.0176 | 10.3481 | 11.7817 | 6.9202 | 8.9445 |
| PRKCSH | 10.8182 | 10.8323 | 11.52 | 9.6587 | 9.158 |
| DHCR24 | 11.119 | 10.4363 | 10.8662 | 5.5813 | 7.6186 |
| S100A10 | 11.9304 | 12.0301 | 11.9165 | 7.1573 | 10.5776 |
| HDGF | 11.3564 | 11.8211 | 12.0185 | 9.0885 | 10.3516 |
| ATP6V0C | 9.6525 | 9.899 | 10.1231 | 7.9139 | 9.0366 |
| SERPING1 | 5.7459 | 5.4992 | 6.0329 | 3.9925 | 3.7251 |
| CYC1 | 11.7322 | 12.0988 | 12.1574 | 6.5572 | 9.1067 |
| POLD2 | 9.1476 | 9.357 | 9.9454 | 5.8966 | 8.2413 |
| PSMC3 | 10.698 | 11.2869 | 11.3805 | 7.1986 | 9.6446 |
| ZFP36L2 | 11.7929 | 12.4243 | 12.194 | 10.9075 | 9.681 |
| TRAM1 | 7.8278 | 7.585 | 8.1262 | 5.7635 | 6.6562 |
| LAMP1 | 8.5708 | 8.1202 | 8.4221 | 6.7776 | 6.8947 |
| NBL1 | 7.4644 | 7.2755 | 7.5796 | 4.7319 | 6.2878 |
| COX6C | 14.1292 | 14.2154 | 14.9972 | 12.4424 | 12.8452 |
| RELA | 8.1625 | 7.8378 | 8.5586 | 6.1343 | 7.1179 |
| SH3GL1 | 7.3366 | 7.7465 | 7.8545 | 5.8716 | 6.7186 |
| CEACAM5 | 9.389 | 8.6775 | 8.3797 | 6.3677 | 7.7386 |
| EPHX1 | 8.4764 | 8.4068 | 8.3899 | 4.9402 | 7.3682 |
| SDC4 | 11.1672 | 11.6725 | 12.9293 | 5.9527 | 10.0088 |
| CHMP2A | 9.3966 | 9.1212 | 8.8505 | 6.3233 | 8.0953 |
| LY6E | 10.7361 | 10.9075 | 11.7568 | 5.2738 | 8.663 |
| PMF1 | 10.2371 | 10.3986 | 10.2524 | 8.8327 | 9.2629 |
| GTF2F1 | 8.6188 | 8.4948 | 9.1645 | 6.2524 | 7.7112 |
| PRPF31 | 9.3173 | 9.8881 | 9.6647 | 8.2126 | 8.353 |
| DHRS3 | 8.2605 | 8.2734 | 8.6338 | 4.2298 | 7.097 |
| VAMP8 | 10.438 | 10.0493 | 10.1944 | 6.2804 | 8.9403 |
| GUSB | 10.7967 | 11.6002 | 12.0013 | 5.5614 | 9.5722 |
| MEN1 | 8.7037 | 8.2318 | 8.6947 | 6.2864 | 7.6035 |
| SNRPD1 | 12.0427 | 11.7639 | 12.0689 | 10.7268 | 11.0151 |
| ITGB2 | 3.8262 | 4.1709 | 4.4497 | 3.0244 | 2.7493 |
| SLPI | 4.9879 | 5.1446 | 4.9761 | 1.7394 | 3.886 |
| EEF1D | 12.0849 | 11.9483 | 11.9994 | 6.4115 | 8.1439 |
| NOTCH3 | 5.8736 | 6.1715 | 6.1086 | 5.1174 | 3.9898 |
| CNOT3 | 9.2442 | 9.757 | 10.0873 | 8.0383 | 8.6062 |
| PPL | 9.1749 | 9.3627 | 9.7934 | 7.2805 | 7.8965 |
| MET | 6.9163 | 6.0288 | 5.6464 | 3.5567 | 4.4341 |
| DGAT1 | 7.6436 | 7.532 | 7.59 | 6.1195 | 5.3076 |
| JUND | 12.2596 | 12.1475 | 12.7646 | 10.5592 | 10.0487 |
| RFC4 | 10.5353 | 10.5015 | 10.8921 | 8.8784 | 9.7102 |
| DCTN3 | 11.9345 | 11.6254 | 11.3717 | 9.7263 | 10.5353 |
| KHSRP | 9.1962 | 9.7866 | 9.9161 | 8.3969 | 8.404 |
| APOC1 | 4.925 | 4.6619 | 5.935 | 1.9993 | 3.921 |
| PARVB | 7.6458 | 7.4091 | 6.7913 | 4.995 | 6.1999 |
| DSC2 | 4.3103 | 4.1177 | 4.4809 | 2.855 | 3.2464 |
| SMAD7 | 5.7806 | 5.4127 | 5.8105 | 4.0167 | 4.7389 |
| PFN2 | 12.1475 | 12.1721 | 11.8831 | 10.1014 | 11.0182 |
| ZNF7 | 7.157 | 7.3472 | 6.8557 | 5.1678 | 6.0371 |
| INHBB | 11.6058 | 11.6972 | 11.1776 | 9.5586 | 9.2071 |
| PFDN4 | 15 | 14.3965 | 14.7995 | 12.9355 | 13.2828 |
| UQCRB | 12.4806 | 12.7369 | 12.5327 | 10.692 | 11.5849 |
| PI4KA | 8.8274 | 9.3696 | 9.3016 | 6.3594 | 7.774 |
| GIPC1 | 9.5226 | 9.4976 | 9.9869 | 8.2413 | 8.6815 |
| TNFRSF1A | 10.9027 | 10.9475 | 11.2148 | 8.9295 | 10.1029 |
| BTBD2 | 8.4611 | 8.5558 | 8.8031 | 6.0202 | 7.3099 |
| CAPN2 | 10.0992 | 10.6146 | 10.0426 | 7.9946 | 9.1297 |
| ELOVL5 | 10.3774 | 9.4911 | 9.0551 | 6.7567 | 8.3339 |
| TAPBP | 8.0035 | 7.7458 | 8.4959 | 6.7694 | 6.844 |
| CYFIP1 | 6.4222 | 6.231 | 6.3926 | 5.4609 | 4.8569 |
| SEC62 | 11.091 | 11.9093 | 11.9686 | 10.2297 | 10.21 |
| CRIP2 | 8.2111 | 8.7114 | 8.4077 | 4.3604 | 6.5342 |
| SF3B4 | 10.529 | 10.4867 | 11.0363 | 8.7261 | 9.4181 |
| MPDU1 | 10.5416 | 10.9801 | 11.0822 | 7.8868 | 9.6321 |
| CLN3 | 8.803 | 8.6061 | 8.4284 | 7.101 | 7.7585 |
| CDT1 | 10.952 | 10.8652 | 10.6401 | 9.0055 | 9.6248 |
| SLC25A1 | 12.9654 | 13.7202 | 13.4528 | 9.3463 | 11.7192 |
| BANF1 | 12.0807 | 12.6561 | 13.1173 | 7.0735 | 10.7221 |
| NUDC | 11.4223 | 11.1051 | 11.5731 | 9.6992 | 10.2247 |
| ARID1A | 8.8585 | 9.4011 | 9.872 | 6.8106 | 8.075 |
| ELF3 | 7.7039 | 7.4211 | 8.0104 | 6.035 | 6.4723 |
| MIOS | 6.7524 | 6.535 | 7.2415 | 4.8633 | 5.6649 |
| PTOV1 | 9.9752 | 9.7922 | 10.3713 | 6.609 | 8.8878 |
| LMNA | 8.5724 | 8.2494 | 8.2727 | 6.7592 | 7.3048 |
| GRINA | 9.687 | 9.6119 | 10.3534 | 5.7364 | 8.0411 |
| INTS1 | 8.7682 | 8.62 | 9.0314 | 7.3422 | 7.5781 |
| ATP6V0A1 | 7.9272 | 7.7158 | 8.3233 | 5.6367 | 6.9517 |
| AVL9 | 7.796 | 8.1364 | 8.0933 | 6.1828 | 7.0221 |
| SCRIB | 10.0984 | 10.3979 | 9.9524 | 6.2705 | 7.5782 |
| BOP1 | 8.8689 | 9.1035 | 9.7091 | 5.7521 | 6.6377 |
| AUTS2 | 1.9083 | 3.4219 | 1.9208 | 0 | 0.2671 |
| EXOC3 | 10.0717 | 9.7611 | 9.3415 | 8.0294 | 8.6772 |
| PPP1R14B | 10.2071 | 10.2808 | 10.9872 | 6.3804 | 9.0836 |
| CIT | 7.9807 | 7.3211 | 7.5353 | 6.06 | 6.5766 |
| RPS6KA2 | 6.6813 | 6.503 | 6.2 | 4.1398 | 4.8081 |
| ZC3HAV1 | 7.682 | 8.157 | 8.3628 | 3.8528 | 6.683 |
| AGO2 | 7.2371 | 7.8713 | 7.3581 | 5.7061 | 6.4612 |
| MINK1 | 9.4496 | 8.9224 | 8.9989 | 6.9616 | 8.0355 |
| GGCT | 11.3638 | 11.8497 | 11.2526 | 10.4528 | 9.9502 |
| GPAA1 | 9.9791 | 9.5155 | 9.7479 | 5.8349 | 6.1798 |
| TSC2 | 8.8363 | 9.3797 | 9.2629 | 5.1822 | 7.4889 |
| EIF2AK1 | 7.4142 | 7.4304 | 7.2564 | 5.3006 | 5.376 |
| NDUFA4 | 11.8717 | 10.2559 | 10.9901 | 6.3259 | 9.4525 |
| PMEPA1 | 7.893 | 8.1367 | 9.3664 | 5.7176 | 6.2934 |
| TMEM214 | 8.3109 | 8.3835 | 9.405 | 5.428 | 7.2147 |
| C19orf53 | 11.7097 | 12.1187 | 12.0381 | 9.5715 | 11.0451 |
| VPS51 | 9.5147 | 9.3515 | 9.7621 | 5.1809 | 6.969 |
| PPDPF | 12.1424 | 12.1977 | 12.1171 | 7.4149 | 10.4274 |
| FXYD5 | 6.6832 | 7.0488 | 6.4403 | 3.896 | 5.6424 |
| RBM22 | 8.9713 | 9.3371 | 9.1537 | 7.613 | 8.0728 |
| SLC52A2 | 8.2913 | 8.5676 | 8.4128 | 6.7081 | 6.6612 |
| APH1A | 11.5952 | 12.3314 | 11.8238 | 9.2874 | 10.8243 |
| SFXN1 | 9.9911 | 9.751 | 10.8568 | 7.5387 | 9.0232 |
| DNAAF5 | 10.3031 | 9.703 | 10.0842 | 7.6036 | 8.7972 |
| BBS1 | 6.8232 | 6.3906 | 6.647 | 5.6454 | 5.3852 |
| COLGALT1 | 8.9433 | 9.5917 | 9.6712 | 6.9309 | 8.4206 |
| NDUFA3 | 11.2374 | 10.674 | 10.6585 | 9.1918 | 9.7692 |
| MOSPD3 | 8.0874 | 8.0653 | 8.5708 | 5.9634 | 7.2931 |
| HGH1 | 7.9199 | 7.7747 | 7.9497 | 6.8668 | 6.4777 |
| RPL36 | 10.2812 | 10.0487 | 10.3725 | 9.0957 | 9.2935 |
| SLCO4A1 | 9.1291 | 9.2104 | 9.1909 | 6.8693 | 7.7087 |
| MOCOS | 7.3994 | 7.8941 | 7.4696 | 5.741 | 6.6546 |
| ZDHHC4 | 8.7006 | 8.861 | 9.3264 | 4.5497 | 7.3595 |
| ZNF580 | 10.9815 | 10.985 | 10.9746 | 7.8941 | 9.5562 |
| SHARPIN | 8.6435 | 8.8784 | 8.7125 | 6.8155 | 7.7122 |
| DBNDD1 | 8.3085 | 8.0355 | 8.1648 | 6.6502 | 7.3406 |
| CPSF1 | 8.5273 | 8.496 | 8.5932 | 5.0709 | 5.7799 |
| SFTPB | 4.9076 | 2.9445 | 4.3653 | 0 | 1.7234 |
| SF3A2 | 8.7612 | 9.0396 | 9.1181 | 5.8256 | 7.7283 |
| CLCN7 | 9.239 | 9.4792 | 8.9518 | 7.9204 | 8.2771 |
| OTUB1 | 11.2243 | 11.5534 | 12.114 | 9.8194 | 10.0541 |
| EHD2 | 8.9713 | 8.5958 | 9.0966 | 6.1586 | 7.8653 |
| CORO1B | 11.4721 | 11.5952 | 11.6396 | 9.6468 | 10.7387 |
| HEATR6 | 10.2297 | 10.0153 | 9.9315 | 8.8451 | 9.1225 |
| VWF | 8.1461 | 7.4527 | 6.4433 | 5.2737 | 5.6434 |
| HSF1 | 9.7598 | 10.5576 | 10.1173 | 6.8452 | 7.8033 |
| C1QB | 9.1411 | 8.4837 | 8.3272 | 3.749 | 6.6967 |
| CLDN3 | 13.1173 | 13.76 | 11.9581 | 11.3113 | 10.7623 |
| RARG | 7.7723 | 8.0844 | 7.8043 | 6.65 | 6.9227 |
| ALOX5 | 6.1766 | 6.9748 | 5.9538 | 4.5529 | 5.0873 |
| C11orf80 | 8.5259 | 8.551 | 8.6009 | 6.3936 | 7.2049 |
| GGCX | 7.8465 | 8.1495 | 7.7644 | 4.47 | 6.7162 |
| C1QL1 | 6.8152 | 6.9001 | 6.8566 | 5.5153 | 5.8047 |
| DSC3 | 7.0792 | 6.6465 | 7.1144 | 5.5055 | 5.6963 |
| LEFTY1 | 7.3789 | 8.4599 | 7.6837 | 6.4011 | 6.442 |
| PTP4A3 | 10.3644 | 9.0688 | 7.9758 | 6.9296 | 6.5029 |
RARG transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network