HCFC1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1272
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: anagrelide
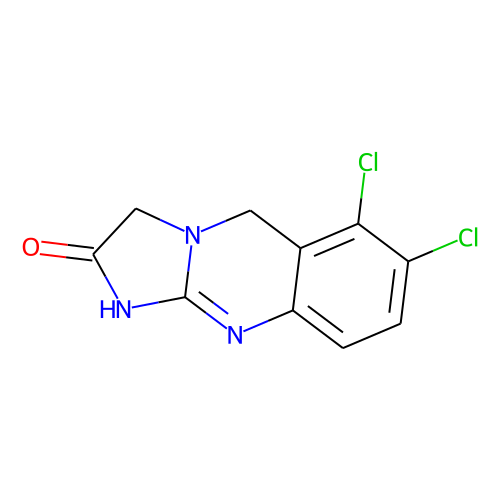
| ID | BRD-K62200014 |
| name | anagrelide |
| type | Small molecule |
| molecular formula | C10H7Cl2N3O |
| molecular weight | 256.08 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | Clc1ccc2N=C3NC(=O)CN3Cc2c1Cl |t:5| |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: HCFC1
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
PDE3A 
|
hsa04924 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| FCGR2C | 5.9785 | 6.0151 | 5.4302 | 4.445 | 4.7798 |
| CLTC | 11.9766 | 12.3268 | 11.9105 | 11.0682 | 11.1912 |
| WIPF2 | 7.1944 | 6.8491 | 6.7711 | 5.7767 | 5.9474 |
| PXN | 11.7483 | 11.9648 | 11.7304 | 10.959 | 10.6172 |
| KTN1 | 10.0611 | 10.374 | 9.3319 | 8.3061 | 8.1266 |
| TIMELESS | 10.5071 | 10.7076 | 10.914 | 9.703 | 9.5637 |
| CAPN1 | 7.0248 | 6.5603 | 7.0441 | 5.275 | 5.4165 |
| SMARCA4 | 7.2081 | 7.0522 | 7.4589 | 5.275 | 6.2975 |
| ICMT | 8.3221 | 8.3384 | 8.7363 | 7.3537 | 7.364 |
| GALE | 9.5997 | 9.2014 | 9.3976 | 7.6253 | 8.0806 |
| USP7 | 7.1591 | 7.228 | 7.3702 | 5.4704 | 6.3243 |
| GPATCH8 | 5.6032 | 5.4963 | 5.4829 | 4.3011 | 4.5243 |
| KLHL21 | 4.196 | 4.0511 | 3.965 | 3.2464 | 3.0634 |
| TARBP1 | 8.204 | 7.8656 | 7.6841 | 6.4579 | 4.541 |
| BAG3 | 12.9606 | 13.054 | 13.0903 | 11.8261 | 12.1058 |
| GNA11 | 7.2399 | 7.4682 | 7.5441 | 5.8683 | 6.4834 |
| RABGGTA | 8.1162 | 8.2048 | 8.7228 | 7.1653 | 7.3855 |
| CAPNS1 | 10.3182 | 9.981 | 10.8824 | 9.0912 | 8.4529 |
| GUK1 | 10.9872 | 10.6526 | 10.6219 | 9.8297 | 9.882 |
| COX4I1 | 13.0973 | 12.825 | 12.9194 | 12.0941 | 11.8831 |
| TOMM20 | 8.8484 | 9.0984 | 9.7033 | 7.35 | 7.972 |
| SRSF11 | 10.0344 | 10.0892 | 9.6995 | 9.0475 | 8.6512 |
| NUMA1 | 7.9544 | 8.2541 | 8.3289 | 7.035 | 7.1121 |
| S100A10 | 11.9304 | 12.0301 | 11.9165 | 10.0665 | 10.9333 |
| RERE | 6.6164 | 6.8513 | 6.5731 | 5.5308 | 5.9297 |
| SYNGR2 | 12.4215 | 12.608 | 11.7084 | 10.9214 | 10.4491 |
| PGD | 7.5012 | 6.5366 | 7.4231 | 5.5618 | 5.7592 |
| IFI30 | 7.778 | 7.906 | 8.2861 | 6.4707 | 6.9594 |
| PRKAB1 | 8.5619 | 8.4774 | 8.851 | 7.3786 | 7.602 |
| CIB1 | 9.8454 | 9.1988 | 9.5886 | 8.2852 | 8.1077 |
| DPF2 | 7.3738 | 7.198 | 7.4026 | 6.1518 | 6.5028 |
| PARP4 | 8.7564 | 8.1821 | 8.9004 | 6.5854 | 7.2987 |
| PRPF3 | 8.6791 | 8.6184 | 8.796 | 7.7639 | 7.9 |
| CTSH | 9.2133 | 7.9626 | 8.6869 | 6.0137 | 6.0481 |
| PSMD12 | 13.054 | 12.759 | 12.6165 | 11.2804 | 11.7704 |
| CSNK1G2 | 8.533 | 8.9426 | 9.0701 | 7.7537 | 7.275 |
| PIEZO1 | 10.5366 | 10.952 | 11.1139 | 8.765 | 9.3139 |
| PIAS3 | 8.3291 | 8.4465 | 8.2788 | 7.4733 | 7.3313 |
| CHSY1 | 7.5345 | 6.7789 | 6.6641 | 4.9364 | 5.8698 |
| GPRC5A | 9.3713 | 9.6671 | 9.1736 | 7.4597 | 7.091 |
| MMD | 6.6649 | 6.2954 | 6.5815 | 4.2593 | 5.4664 |
| SUCO | 10.2888 | 9.9115 | 9.8314 | 8.3802 | 8.1183 |
| COIL | 10.2724 | 9.9727 | 10.1917 | 9.1744 | 8.8659 |
| LLGL2 | 8.0172 | 7.735 | 8.3176 | 6.732 | 4.7567 |
| TDG | 11.4997 | 11.6483 | 11.1411 | 10.2897 | 9.9853 |
| CD151 | 7.5885 | 7.3497 | 8.3418 | 6.1141 | 6.5149 |
| KIAA0513 | 5.7992 | 5.7246 | 5.6187 | 3.8192 | 4.472 |
| KIF23 | 8.9106 | 9.0408 | 9.0195 | 8.0688 | 8.2331 |
| KRT15 | 9.527 | 9.3816 | 8.4149 | 6.4862 | 7.4843 |
| NME3 | 12.8372 | 12.6102 | 12.4731 | 11.0042 | 10.7193 |
| EMP2 | 11.4864 | 11.7558 | 10.6116 | 8.9897 | 9.895 |
| PITRM1 | 8.4821 | 7.4211 | 8.8958 | 6.4086 | 6.4446 |
| TCN1 | 6.0041 | 8.5503 | 9.1985 | 3.9725 | 3.2909 |
| ALDH3B1 | 5.8861 | 5.8735 | 6.5498 | 4.7195 | 5.061 |
| HOXC6 | 4.5386 | 3.804 | 5.761 | 2.296 | 2.5953 |
| TMED3 | 7.51 | 7.862 | 8.2294 | 6.4811 | 6.8994 |
| NADK | 9.0683 | 8.712 | 8.8757 | 7.9826 | 7.813 |
| LGALS3 | 11.2822 | 12.0871 | 12.7493 | 10.0077 | 10.468 |
| FADS1 | 10.4687 | 11.2062 | 10.6623 | 8.8996 | 9.6621 |
| KDM2A | 7.2968 | 7.0269 | 7.2049 | 6.4115 | 6.0673 |
| AKR1C2 | 7.4673 | 6.6377 | 6.8607 | 4.5721 | 4.4344 |
| TM7SF2 | 7.4511 | 7.2765 | 7.2416 | 6.1354 | 6.5202 |
| CROCCP2 | 7.143 | 7.1703 | 8.005 | 6.1142 | 5.5101 |
| EIF4B | 9.8153 | 9.1084 | 9.6622 | 8.2825 | 7.7316 |
| RBM25 | 9.2989 | 9.2219 | 9.2908 | 8.3566 | 8.0169 |
| NIPA2 | 8.0061 | 7.3872 | 7.823 | 6.4072 | 6.5002 |
| FNBP4 | 8.3201 | 8.4268 | 8.4792 | 7.1433 | 7.5827 |
| KDM1A | 9.5301 | 9.7775 | 10.044 | 8.7435 | 8.7756 |
| CDK11A | 9.3611 | 8.8365 | 9.2982 | 8.0157 | 8.0567 |
| NME4 | 9.8662 | 9.9766 | 10.4805 | 8.8148 | 7.4546 |
| CEMIP | 5.3156 | 5.2934 | 5.1222 | 3.6571 | 4.3782 |
| ANXA2 | 14.3237 | 15 | 15 | 13.4927 | 13.6894 |
| KLC1 | 7.8899 | 7.5984 | 7.4343 | 6.6032 | 6.402 |
| PCGF2 | 8.0899 | 7.333 | 8.1274 | 6.0596 | 6.0839 |
| HNRNPA1 | 11.8602 | 12.5616 | 11.9304 | 10.8811 | 10.9123 |
| CDC42EP4 | 8.9031 | 9.4343 | 9.3075 | 7.7122 | 7.1254 |
| AKR1C1 | 7.8868 | 7.5161 | 7.3027 | 5.7235 | 5.0476 |
| NPLOC4 | 8.4721 | 8.061 | 8.333 | 6.9074 | 7.0866 |
| YTHDF2 | 8.209 | 7.9944 | 8.3167 | 6.0681 | 7.1534 |
| SLTM | 7.7458 | 8.3062 | 7.7225 | 6.9232 | 6.7121 |
| VPS51 | 9.5147 | 9.3515 | 9.7621 | 8.2409 | 7.4195 |
| FN3KRP | 9.2691 | 8.9927 | 8.919 | 8.1459 | 7.8379 |
| ATG101 | 8.9882 | 8.798 | 8.4094 | 7.6864 | 7.7403 |
| ZC3H7A | 8.3983 | 7.849 | 8.1715 | 6.844 | 7.1827 |
| KANK2 | 6.3619 | 5.9084 | 6.3025 | 4.2959 | 4.8018 |
| SNRNP25 | 12.2185 | 12.9861 | 12.9606 | 11.4883 | 11.1634 |
| PNPO | 9.8654 | 9.8198 | 9.6145 | 8.923 | 8.3284 |
| ECHDC2 | 6.6515 | 6.3638 | 6.4982 | 5.2432 | 4.2593 |
| YRDC | 6.2854 | 6.6867 | 6.8788 | 5.556 | 5.4319 |
| PITPNC1 | 9.2844 | 8.2947 | 8.8991 | 7.6169 | 7.2397 |
| LTBP3 | 6.2392 | 5.6143 | 6.9603 | 4.7878 | 3.8471 |
| MOCOS | 7.3994 | 7.8941 | 7.4696 | 5.9251 | 6.0097 |
| USP48 | 6.7917 | 6.5583 | 7.4586 | 5.7474 | 5.1296 |
| GDF15 | 13.8087 | 14.3437 | 13.8087 | 10.7873 | 12.0152 |
| LDLRAP1 | 7.1205 | 7.091 | 6.881 | 5.8455 | 5.715 |
| TBC1D16 | 7.5184 | 7.6596 | 7.728 | 6.6281 | 6.6216 |
| LRRC59 | 9.6944 | 9.8788 | 10.0435 | 8.7974 | 7.3735 |
| ARAP1 | 7.3448 | 7.7202 | 7.2942 | 6.479 | 6.5654 |
| SPSB3 | 8.9982 | 9.293 | 9.1783 | 8.3291 | 7.5651 |
| OXLD1 | 10.9732 | 10.7451 | 10.8871 | 9.9967 | 10.0541 |
| CORO1B | 11.4721 | 11.5952 | 11.6396 | 10.2772 | 10.3326 |
| MTA2 | 6.3142 | 6.0255 | 6.3049 | 5.2014 | 5.0833 |
| HBZ | 5.9056 | 6.0876 | 5.8906 | 4.7911 | 4.5097 |
| SAA4 | 6.0569 | 5.5295 | 6.0192 | 4.9763 | 4.3182 |
HCFC1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: HCFC1
[2] Insulin Receptor Associates with Promoters Genome-wide and Regulates Gene Expression.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below