NCAPG2 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1276
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: milrinone
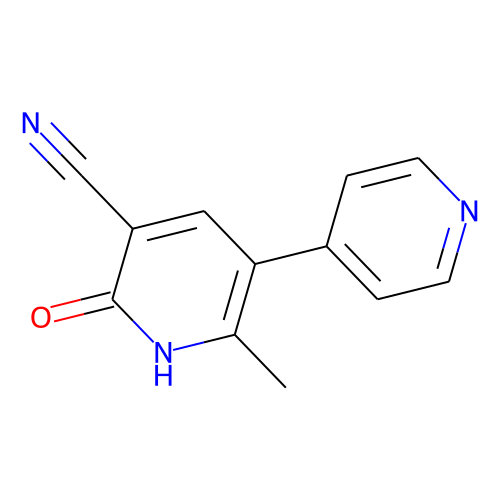
| ID | BRD-K67080878 |
| name | milrinone |
| type | Small molecule |
| molecular formula | C12H9N3O |
| molecular weight | 211.22 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | Cc1[nH]c(=O)c(cc1-c1ccncc1)C#N |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: NCAPG2
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
PDE2A 
|
hsa05032 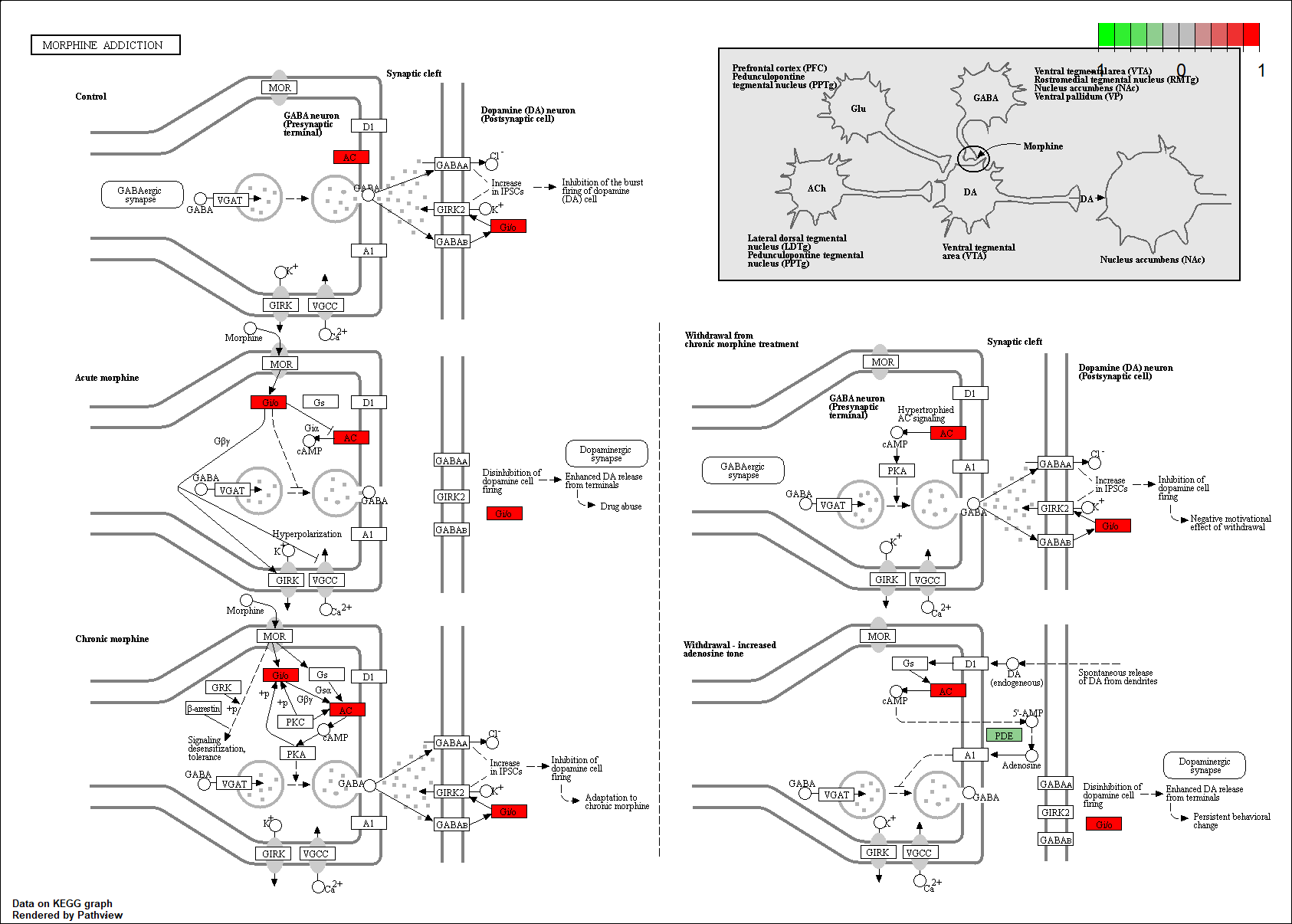
|
|
PDE3A 
|
hsa04022 
|
|
PDE3B 
|
hsa05032 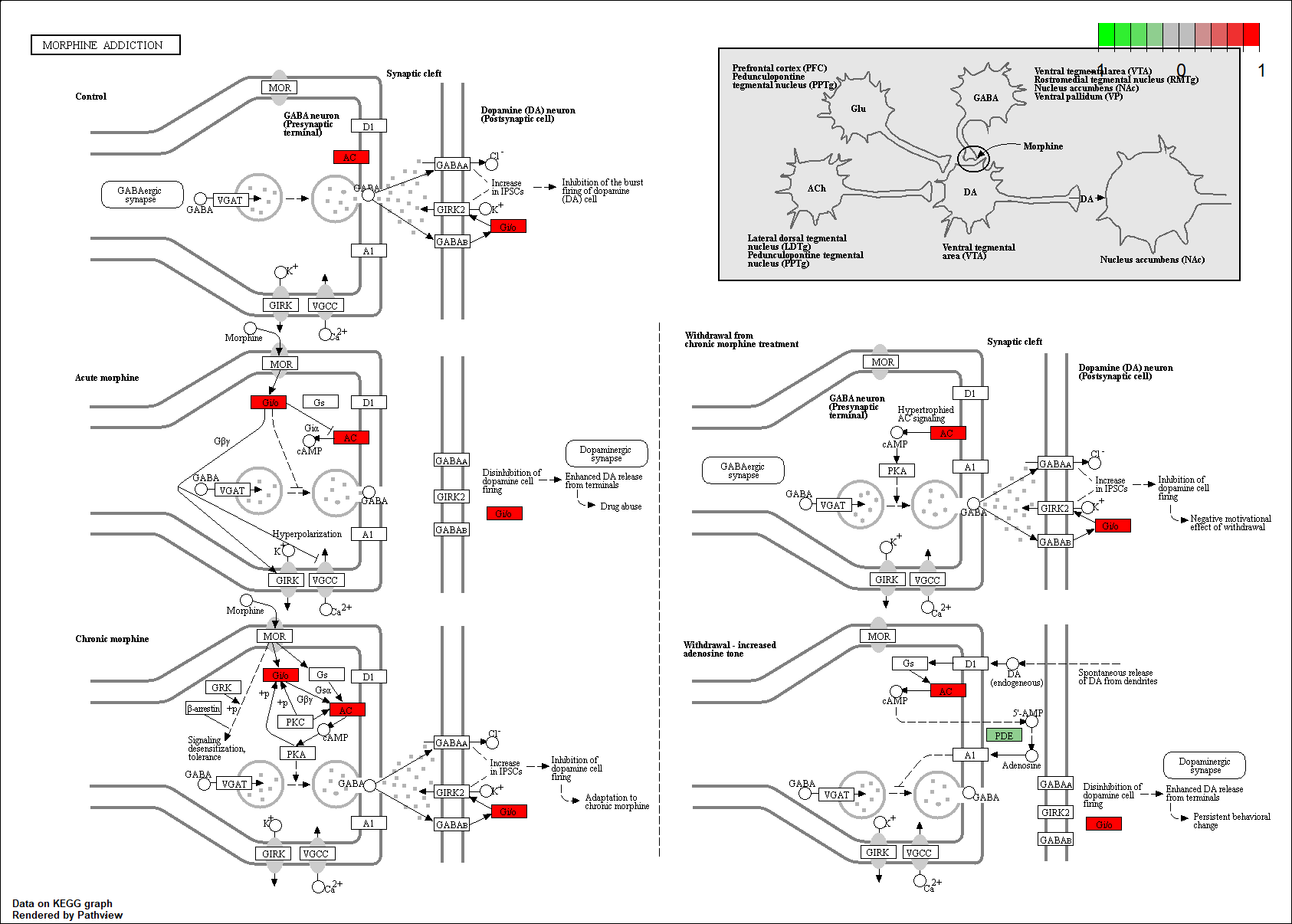
|
|
PDE5A 
|
hsa00230 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| FCGR2B | 6.1602 | 5.8933 | 5.997 | 4.3935 | 5.0641 |
| GTF2H2B | 7.3289 | 7.8504 | 7.2922 | 5.7525 | 6.1318 |
| ATP6V0A4 | 7.5461 | 7.2523 | 7.0926 | 6.258 | 6.3051 |
| LRRC31 | 7.1443 | 6.9685 | 6.4555 | 5.9965 | 5.1861 |
| SIKE1 | 9.8549 | 9.6299 | 10.2997 | 8.9943 | 8.9452 |
| TCL1A | 6.7754 | 6.4441 | 7.1464 | 5.6548 | 5.788 |
| TJAP1 | 8.6281 | 8.8647 | 8.4235 | 7.761 | 7.8511 |
| RBM12B | 7.7742 | 7.5829 | 8.0118 | 7.118 | 6.3048 |
| MAPKAPK2 | 11.2485 | 9.784 | 11.3783 | 9.0783 | 8.8585 |
| BLCAP | 9.3058 | 9.138 | 9.1829 | 8.4979 | 8.4886 |
| POLG2 | 8.5821 | 8.606 | 8.5723 | 4.5677 | 5.2871 |
| TPD52L2 | 8.811 | 8.773 | 7.6676 | 7.0198 | 7.1212 |
| TIAM1 | 6.099 | 5.9678 | 5.8802 | 5.3339 | 5.2104 |
| CDH3 | 7.7587 | 7.3525 | 7.3644 | 6.8181 | 6.188 |
| ADCY9 | 7.5783 | 6.4484 | 6.4336 | 5.4323 | 5.1793 |
| PRKCSH | 10.8182 | 10.8323 | 11.52 | 10.2291 | 9.7487 |
| LGALS3BP | 7.9395 | 8.4475 | 8.4267 | 5.6008 | 7.1763 |
| ZFP36L2 | 11.7929 | 12.4243 | 12.194 | 11.342 | 10.5285 |
| PTDSS1 | 9.7437 | 9.9329 | 9.7441 | 8.9518 | 7.6783 |
| IFITM1 | 6.6506 | 6.7032 | 6.9171 | 4.6817 | 4.4852 |
| NBL1 | 7.4644 | 7.2755 | 7.5796 | 6.2381 | 6.082 |
| LPCAT1 | 12.6997 | 12.7493 | 13.1812 | 11.4049 | 11.0651 |
| LY6E | 10.7361 | 10.9075 | 11.7568 | 9.18 | 10.0541 |
| VEZF1 | 9.4693 | 9.5593 | 9.0677 | 7.5546 | 8.512 |
| BLVRB | 12.4529 | 12.5855 | 12.9146 | 11.4925 | 11.9002 |
| CYP1B1 | 10.254 | 9.8014 | 9.5145 | 8.6346 | 7.8757 |
| CTSK | 9.8039 | 9.6668 | 8.8092 | 7.2096 | 8.0551 |
| PFKFB3 | 10.1213 | 10.8313 | 10.4373 | 8.6673 | 9.2629 |
| DHRS3 | 8.2605 | 8.2734 | 8.6338 | 7.2977 | 7.2796 |
| PIEZO1 | 10.5366 | 10.952 | 11.1139 | 9.3001 | 9.8921 |
| COIL | 10.2724 | 9.9727 | 10.1917 | 9.3892 | 9.4382 |
| DGAT1 | 7.6436 | 7.532 | 7.59 | 6.2016 | 6.9844 |
| JUND | 12.2596 | 12.1475 | 12.7646 | 10.2951 | 10.9078 |
| MMP11 | 9.0961 | 9.593 | 8.7085 | 6.5111 | 8.1139 |
| STK3 | 8.591 | 8.3202 | 7.718 | 6.9143 | 7.0245 |
| TRIM16 | 9.4056 | 9.1749 | 8.991 | 8.2126 | 8.256 |
| SLC25A24 | 10.5733 | 10.6133 | 10.3031 | 9.4216 | 9.7574 |
| KIAA0513 | 5.7992 | 5.7246 | 5.6187 | 4.3399 | 4.6868 |
| PPM1D | 9.4695 | 10.0892 | 10.2934 | 8.6349 | 8.982 |
| EXO1 | 8.6546 | 8.4742 | 8.4324 | 7.8344 | 7.6521 |
| TFF3 | 9.3276 | 8.7471 | 7.6199 | 6.586 | 6.775 |
| ATP7B | 7.2773 | 7.7278 | 7.6033 | 6.6931 | 6.6483 |
| FOXA1 | 10.1464 | 10.6837 | 9.9109 | 8.4427 | 9.2621 |
| KIF23 | 8.9106 | 9.0408 | 9.0195 | 8.191 | 8.306 |
| TFF1 | 12.3368 | 12.8006 | 12.9943 | 9.0638 | 7.4286 |
| ISG15 | 9.2057 | 9.5334 | 8.8947 | 5.9257 | 8.0344 |
| GREB1 | 7.0646 | 6.6143 | 6.9195 | 5.0153 | 5.8287 |
| CSNK1D | 10.2112 | 9.9865 | 10.3252 | 9.3197 | 9.3297 |
| SF1 | 9.8564 | 9.7023 | 9.9949 | 8.5219 | 9.1284 |
| IRF7 | 8.0703 | 7.8639 | 7.746 | 6.9275 | 5.872 |
| CAPN2 | 10.0992 | 10.6146 | 10.0426 | 8.5864 | 9.4024 |
| MDK | 10.1832 | 11.0664 | 10.6763 | 8.5181 | 9.2732 |
| TNFRSF10B | 8.622 | 8.597 | 9.8438 | 7.1237 | 7.6639 |
| SIN3B | 7.4003 | 7.4295 | 7.1406 | 5.8273 | 6.6428 |
| PHLDA2 | 11.0721 | 11.8336 | 11.3764 | 9.703 | 10.3312 |
| AP2S1 | 12.9774 | 12.5005 | 13.1972 | 11.652 | 12.0363 |
| RPS6KB1 | 11.2836 | 11.478 | 11.5413 | 10.324 | 10.3567 |
| TMPRSS2 | 5.4706 | 5.1678 | 5.0307 | 3.7129 | 4.3145 |
| PXDN | 11.5683 | 13.2307 | 12.2908 | 10.1531 | 10.431 |
| IFITM3 | 11.4939 | 11.483 | 11.7657 | 10.6696 | 9.5901 |
| FASN | 11.845 | 11.8958 | 12.2155 | 10.7782 | 11.309 |
| CDC34 | 10.2621 | 10.2666 | 10.2666 | 8.7701 | 9.0786 |
| CEMIP | 5.3156 | 5.2934 | 5.1222 | 3.5289 | 3.1958 |
| ACKR3 | 10.6952 | 10.696 | 9.9898 | 7.7919 | 8.8958 |
| FANCI | 9.8319 | 9.7336 | 10.0031 | 8.8426 | 9.0566 |
| ADCY1 | 6.5369 | 6.2272 | 6.0605 | 3.4545 | 5.1895 |
| DPY19L4 | 9.4045 | 8.9597 | 9.1151 | 7.9354 | 8.0468 |
| MXRA8 | 5.9343 | 5.8858 | 5.3408 | 4.341 | 4.6038 |
| KLC1 | 7.8899 | 7.5984 | 7.4343 | 6.535 | 6.8876 |
| RSAD2 | 4.8862 | 4.5006 | 4.6957 | 3.3337 | 3.7952 |
| CA12 | 10.023 | 10.5107 | 10.3516 | 9.0785 | 9.0209 |
| GLA | 9.8042 | 10.0913 | 9.9556 | 8.4353 | 8.9529 |
| ARMCX6 | 12.8289 | 11.983 | 11.9267 | 10.4706 | 11.1261 |
| ARHGEF10 | 7.4567 | 6.575 | 7.7048 | 5.703 | 5.5415 |
| C3 | 8.6952 | 9.503 | 9.5812 | 6.8957 | 7.788 |
| PPDPF | 12.1424 | 12.1977 | 12.1171 | 10.9333 | 11.3348 |
| PDXK | 8.1969 | 8.1126 | 8.2194 | 7.249 | 7.4357 |
| TSEN34 | 11.0112 | 11.1129 | 10.9702 | 9.9851 | 10.1832 |
| RMND5B | 9.9661 | 9.515 | 9.7574 | 8.6302 | 8.9391 |
| KANK2 | 6.3619 | 5.9084 | 6.3025 | 3.6082 | 5.2432 |
| UFSP2 | 6.3088 | 6.4023 | 6.0151 | 5.1773 | 5.0984 |
| PITPNC1 | 9.2844 | 8.2947 | 8.8991 | 7.2955 | 7.8559 |
| TIPIN | 8.7702 | 8.3725 | 8.7503 | 7.7223 | 7.9352 |
| PIEZO2 | 6.834 | 5.9818 | 5.939 | 4.7765 | 4.2421 |
| ABHD17A | 9.7581 | 9.5325 | 9.0978 | 8.0977 | 8.3596 |
| TCF25 | 9.6611 | 10.2235 | 10.5675 | 8.1853 | 9.2104 |
| CRISPLD2 | 5.774 | 6.4344 | 5.0491 | 3.1849 | 4.4134 |
| GDF15 | 13.8087 | 14.3437 | 13.8087 | 8.9902 | 11.7322 |
| UGCG | 6.059 | 6.4411 | 7.0657 | 3.1407 | 4.0739 |
| MPHOSPH8 | 6.263 | 6.9221 | 6.3578 | 4.5595 | 5.0565 |
| LDLRAP1 | 7.1205 | 7.091 | 6.881 | 6.0189 | 5.469 |
| THOC2 | 8.4946 | 8.0125 | 8.2194 | 6.5245 | 7.2127 |
| DNPEP | 10.4888 | 10.4627 | 10.5125 | 9.0891 | 9.4306 |
| C11orf24 | 7.3526 | 7.411 | 7.7443 | 6.4182 | 5.9748 |
| OGFOD3 | 10.6002 | 10.7608 | 10.2508 | 9.6752 | 9.4494 |
| AEBP1 | 8.9211 | 8.1924 | 7.816 | 7.2702 | 6.038 |
| GAS6 | 8.473 | 8.0341 | 8.0905 | 6.6498 | 6.9885 |
| SERPINF1 | 7.0345 | 7.3811 | 7.9875 | 3.1807 | 5.2767 |
| CLDN5 | 8.6567 | 7.8035 | 8.312 | 5.8046 | 7.0455 |
| PDE4A | 8.187 | 8.0907 | 8.3095 | 6.9682 | 7.5294 |
| GFRA1 | 6.9863 | 6.8215 | 6.8555 | 6.2204 | 6.016 |
| TRH | 6.0877 | 5.6952 | 6.4814 | 4.6667 | 4.6474 |
NCAPG2 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: NCAPG2
[7] PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression.
[8] Novel significant stage-specific differentially expressed genes in hepatocellular carcinoma.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below