ZNF701 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1396
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: levosulpiride
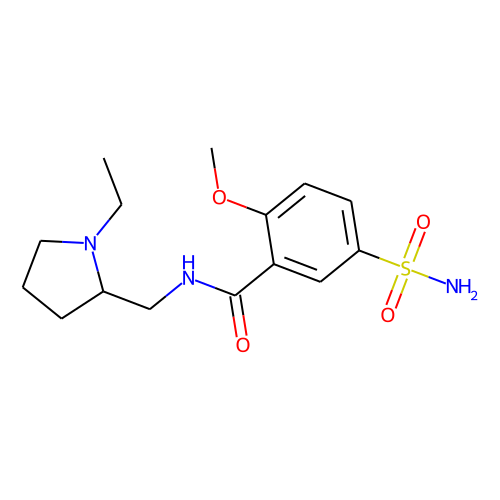
| ID | BRD-K51671335 |
| name | levosulpiride |
| type | Small molecule |
| molecular formula | C15H23N3O4S |
| molecular weight | 341.4 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | CCN1CCC[C@H]1CNC(=O)c1cc(ccc1OC)S(N)(=O)=O |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: ZNF701
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CA12 
|
hsa00910 
|
|
CA1 |
hsa00910 
|
|
CA7 
|
hsa00910 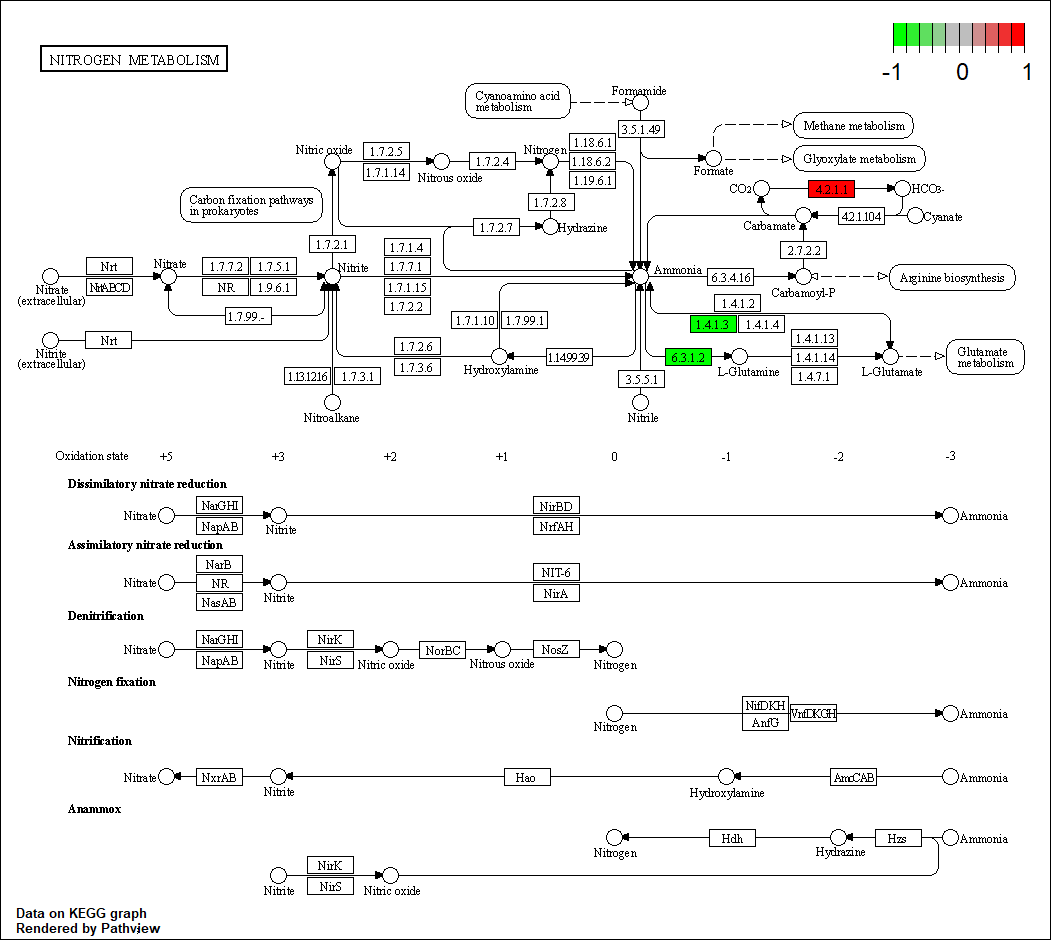
|
|
DRD2 |
|
|
DRD3 
|
hsa04728 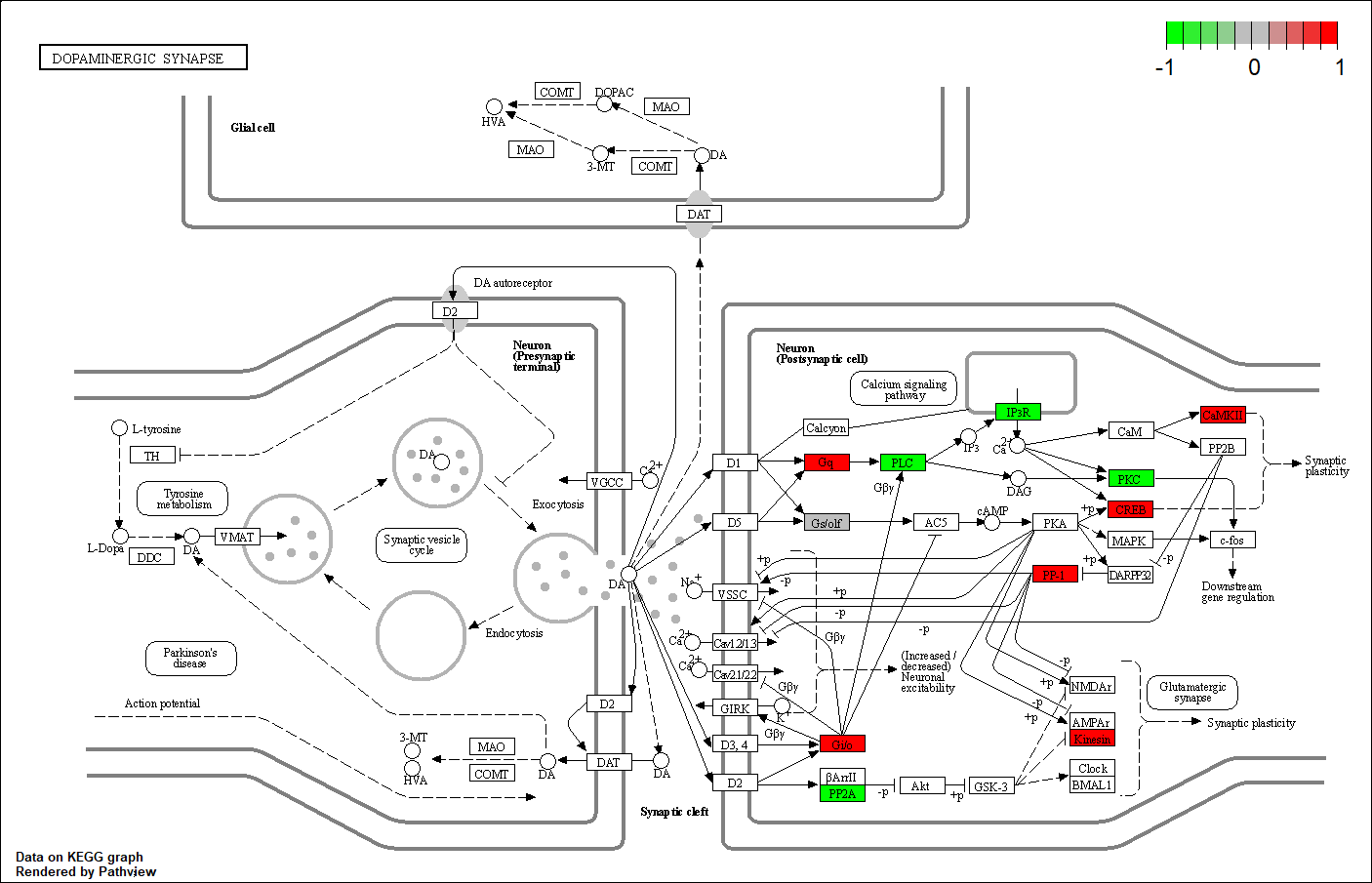
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| CAPN9 | 5.6353 | 6.5721 | 6.078 | 4.0455 | 4.0564 |
| NDRG4 | 7.401 | 7.6978 | 7.8088 | 5.0693 | 5.7455 |
| ENTPD1 | 5.3422 | 6.2311 | 6.3249 | 2.3172 | 3.6857 |
| LDB3 | 7.0269 | 7.2343 | 7.198 | 5.3787 | 5.6005 |
| TBC1D30 | 8.2083 | 8.5905 | 8.2397 | 5.8161 | 6.3904 |
| MYCL | 4.7125 | 4.9784 | 5.0739 | 3.2931 | 3.3927 |
| HHEX | 6.9632 | 6.5834 | 6.0751 | 3.8776 | 4.2903 |
| OTUB2 | 7.6095 | 6.7519 | 6.768 | 4.8118 | 4.7502 |
| CALHM2 | 8.4162 | 7.4806 | 8.1607 | 5.4863 | 5.9231 |
| CDC42 | 11.0858 | 11.7015 | 12.4145 | 8.598 | 9.0175 |
| PPP1R13B | 8.3031 | 7.4018 | 7.1181 | 4.9612 | 5.0519 |
| CHEK1 | 8.2846 | 8.166 | 8.0671 | 6.4543 | 6.5679 |
| MKNK1 | 8.9545 | 8.4191 | 9.6005 | 5.1396 | 4.3994 |
| NUP88 | 10.2421 | 10.7026 | 11.0443 | 8.2257 | 7.7947 |
| KTN1 | 9.4215 | 8.6365 | 8.904 | 5.8488 | 5.55 |
| SCYL3 | 5.3468 | 5.4565 | 5.4763 | 3.2455 | 3.4392 |
| DHRS7 | 9.0672 | 9.5833 | 9.304 | 7.0129 | 6.7942 |
| SLC35F2 | 8.0534 | 7.15 | 6.5498 | 3.6301 | 4.1867 |
| SMAD3 | 8.0334 | 7.6853 | 7.66 | 5.5729 | 5.7759 |
| OXA1L | 13.2629 | 12.3974 | 12.4494 | 4.5233 | 8.6608 |
| CDCA4 | 10.3152 | 9.5681 | 8.9627 | 4.1874 | 5.0893 |
| PLSCR3 | 9.5613 | 8.7626 | 8.3876 | 5.964 | 6.0085 |
| PXMP2 | 11.3608 | 11.0484 | 10.8431 | 8.1147 | 8.4542 |
| CLPX | 9.1834 | 8.4818 | 8.4027 | 6.182 | 5.5751 |
| TXNL4B | 9.1149 | 8.4967 | 8.5734 | 5.2425 | 5.262 |
| TM9SF3 | 7.8558 | 9.4529 | 8.7729 | 4.6981 | 6.2042 |
| PACSIN3 | 9.2435 | 8.391 | 8.2846 | 6.182 | 6.3496 |
| MACF1 | 8.7767 | 8.4101 | 7.7648 | 5.4748 | 4.8884 |
| COG2 | 9.1483 | 9.1914 | 9.6691 | 4.7545 | 4.4703 |
| WASF3 | 7.2226 | 6.2871 | 6.2053 | 3.7905 | 4.3994 |
| RNPS1 | 14.0179 | 14.7898 | 14.2106 | 10.4604 | 10.1528 |
| DENND2D | 9.1483 | 8.2122 | 8.38 | 5.2425 | 5.121 |
| ABHD4 | 7.5579 | 6.3578 | 6.7522 | 3.7905 | 4.1589 |
| RPP38 | 9.3353 | 9.6667 | 8.4242 | 6.0228 | 5.8542 |
| CIAPIN1 | 10.614 | 9.8286 | 8.7624 | 6.182 | 6.2905 |
| COG7 | 9.3464 | 9.0542 | 9.4556 | 5.8575 | 5.3929 |
| HACD1 | 7.6532 | 7.6904 | 7.6174 | 6.0365 | 6.0781 |
| MARCKSL1 | 11.1455 | 12.0814 | 11.9439 | 8.5457 | 9.548 |
| WSB2 | 11.0581 | 11.2996 | 11.3604 | 9.6423 | 8.2968 |
| ARIH1 | 7.5459 | 7.6452 | 7.5659 | 5.8373 | 6.2283 |
| CDK2AP1 | 13.4713 | 13.4506 | 12.5927 | 9.9165 | 8.5077 |
| RNF41 | 7.9383 | 8.4093 | 8.0187 | 6.5066 | 6.6097 |
| KIF5B | 9.7524 | 10.0345 | 10.2104 | 7.5206 | 6.4268 |
| CTSH | 7.0776 | 8.7087 | 8.8004 | 4.2687 | 3.2908 |
| CTSK | 7.1327 | 7.1515 | 7.3372 | 5.1041 | 3.6548 |
| EIF2B2 | 9.33 | 9.8937 | 9.6507 | 8.0086 | 7.7767 |
| ARL2 | 9.1786 | 9.2385 | 9.3525 | 7.4842 | 6.8837 |
| SRSF5 | 10.096 | 9.6861 | 9.3207 | 7.9357 | 7.7975 |
| STAM | 7.9377 | 7.7171 | 8.2838 | 3.1132 | 3.5124 |
| TOE1 | 7.931 | 7.5748 | 7.4823 | 6.1509 | 6.0087 |
| F3 | 5.396 | 7.0626 | 5.8245 | 2.6693 | 2.6104 |
| ATG14 | 8.5121 | 8.3055 | 9.0572 | 6.0924 | 6.5817 |
| IDI1 | 10.5901 | 11.1134 | 10.4329 | 8.0942 | 8.3208 |
| ISG15 | 8.6481 | 9.1096 | 10.0173 | 4.6312 | 6.6077 |
| TCN1 | 9.4396 | 7.893 | 7.668 | 1.7004 | 4.7322 |
| ARHGAP12 | 6.0428 | 6.9843 | 7.0122 | 4.0503 | 4.1255 |
| SLC19A2 | 9.4761 | 9.4142 | 8.9672 | 6.3215 | 7.1396 |
| UCK2 | 10.7786 | 10.6912 | 11.0622 | 9.0675 | 9.1197 |
| APEX1 | 13.3267 | 12.778 | 13.4808 | 10.4111 | 10.2247 |
| SH3GLB1 | 6.3548 | 6.658 | 6.5874 | 5.1551 | 4.7898 |
| USP15 | 8.0143 | 8.3272 | 8.9769 | 5.5591 | 5.8874 |
| RSL1D1 | 10.8003 | 11.5703 | 11.8375 | 8.2434 | 8.1283 |
| MOAP1 | 10.0566 | 10.7018 | 10.2941 | 5.0485 | 7.0361 |
| SAMD4A | 12.2136 | 12.9974 | 12.5135 | 8.2786 | 9.8014 |
| KLC1 | 8.444 | 7.4784 | 7.5861 | 4.5258 | 4.7558 |
| MAP3K9 | 7.2327 | 7.4127 | 7.2193 | 5.7046 | 5.8906 |
| POLR2C | 9.8268 | 9.1229 | 9.3069 | 7.8102 | 7.101 |
| MIR22HG | 4.5513 | 5.0981 | 4.4307 | 1.8567 | 2.8143 |
| DLST | 11.9645 | 10.7103 | 11.1814 | 8.5165 | 8.3756 |
| METTL9 | 9.8423 | 10.0566 | 10.4025 | 6.8833 | 6.9626 |
| TPCN1 | 7.5458 | 7.1719 | 6.6752 | 5.3136 | 4.8729 |
| EFHD2 | 9.7479 | 11.2636 | 10.5431 | 7.8308 | 7.8583 |
| POLR3E | 10.0867 | 10.7093 | 9.9472 | 7.5473 | 7.7954 |
| AP5M1 | 8.9471 | 8.7416 | 8.8531 | 5.4423 | 6.9258 |
| DHX32 | 9.0958 | 8.9302 | 9.3379 | 6.0025 | 6.8504 |
| PNMA1 | 10.4853 | 10.2654 | 9.9008 | 7.9624 | 6.4516 |
| GTPBP4 | 12.9649 | 11.0807 | 11.6077 | 8.1194 | 8.7696 |
| ECHDC2 | 5.7086 | 5.4084 | 5.6178 | 3.6356 | 4.0513 |
| CHFR | 10.4713 | 10.1523 | 10.5597 | 8.0236 | 6.3238 |
| C14orf132 | 9.0005 | 9.1775 | 9.0203 | 6.8047 | 5.7401 |
| MTPAP | 10.0145 | 9.2778 | 9.1749 | 6.2729 | 7.0469 |
| MYO5C | 7.3667 | 8.5561 | 7.8105 | 3.052 | 3.7566 |
| NIP7 | 9.2576 | 9.9578 | 10.048 | 6.0632 | 7.0421 |
| RHOF | 9.3772 | 8.4709 | 8.0816 | 5.6311 | 6.2859 |
| HEATR3 | 6.8604 | 7.6358 | 7.4147 | 5.5001 | 5.4577 |
| COA7 | 8.6078 | 8.3048 | 8.2451 | 5.9161 | 6.7558 |
| NXN | 12.5482 | 9.0534 | 10.6626 | 6.3288 | 5.7958 |
| ZFYVE21 | 8.1791 | 8.0921 | 7.896 | 6.5293 | 6.0176 |
| TRIM8 | 7.2066 | 7.6328 | 7.7458 | 3.6946 | 5.3437 |
| USP3 | 9.6345 | 11.1763 | 11.541 | 6.5336 | 7.6404 |
| RAB35 | 9.2381 | 9.1098 | 9.423 | 7.0898 | 7.5212 |
| FAM174B | 7.3668 | 7.7752 | 7.7387 | 5.8743 | 5.1439 |
| HIP1R | 10.5567 | 10.804 | 10.373 | 8.5964 | 8.2685 |
| OTUB1 | 11.6812 | 11.692 | 11.6814 | 10.1222 | 9.817 |
| CTNNBIP1 | 8.4168 | 8.072 | 8.0831 | 6.7401 | 5.7626 |
| UMOD | 4.8165 | 4.2594 | 4.5479 | 3.0631 | 2.4642 |
ZNF701 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: ZNF701
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa05168