PTPN11 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1421
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: varenicline

| ID | BRD-K18855837 |
| name | varenicline |
| type | Small molecule |
| molecular formula | C13H13N3 |
| molecular weight | 211.26 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | "C1[C@H]2CNC[C@@H]1c1cc3nccnc3cc21 |&1:1,5|" |
CONDITION TYPE: trt_cp
Compound
CELL LINE: A549
Cell lines used for experiments
TRANSCRIPTION FACTOR: PTPN11
Transcription regulatory factors corresponding to this network
ARTICLE: varenicline
[1] Smoking Cessation in Chronic Obstructive Pulmonary Disease.
[2] Discovery and development of varenicline for smoking cessation.
[4] SMOKING CESSATION TREATMENTS: CURRENT PSYCHOLOGICAL AND PHARMACOLOGICAL OPTIONS.
[5] [Interventions for smoking cessation in 2018].
[6] Smoking cessation for people with chronic obstructive pulmonary disease.
[7] Pharmacotherapy for Substance Use Disorders.
[8] Combination bupropion SR and varenicline for smoking cessation: a systematic review.
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CHRNA3 |
|
|
CHRNA4 
|
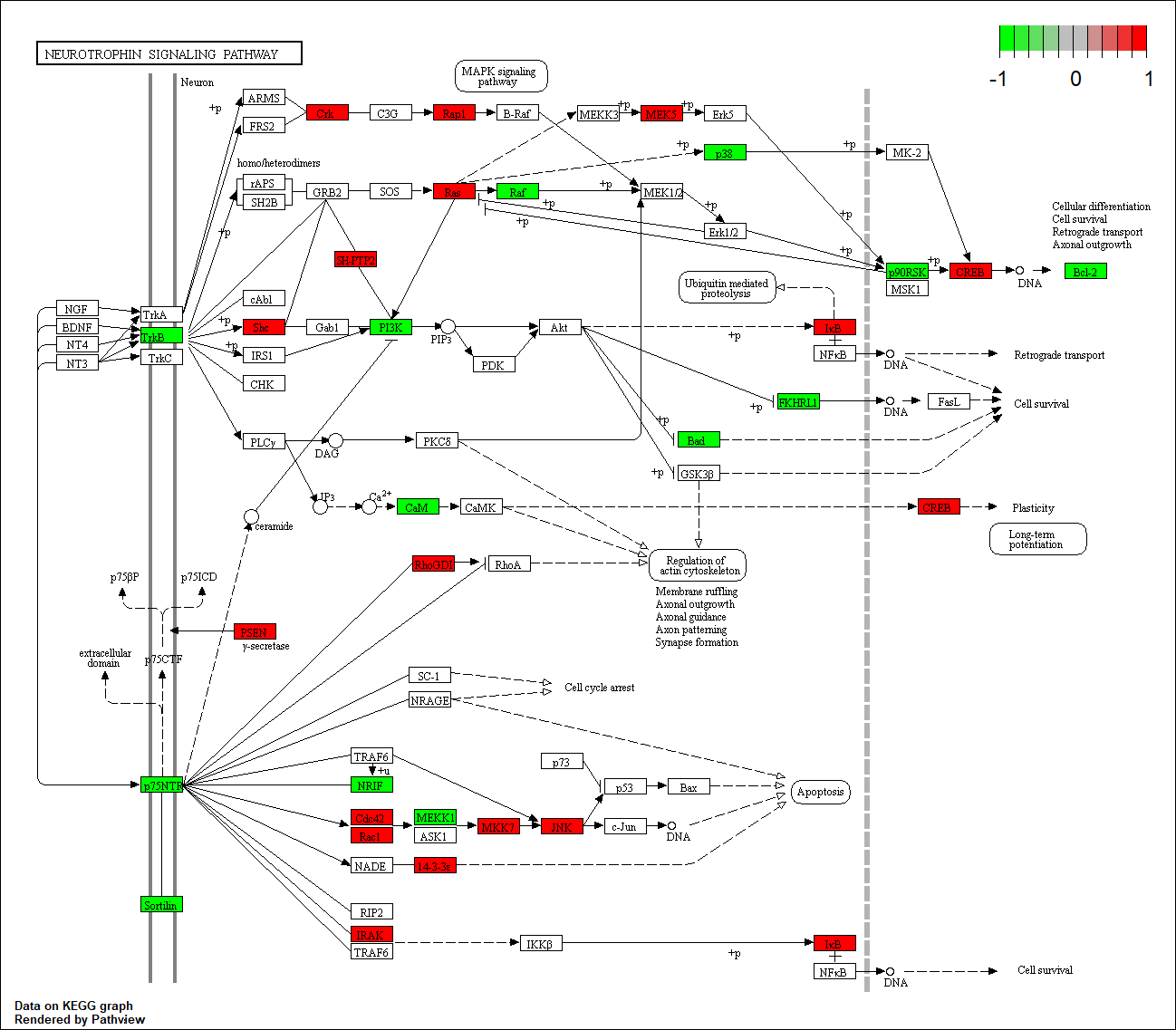
hsa05033 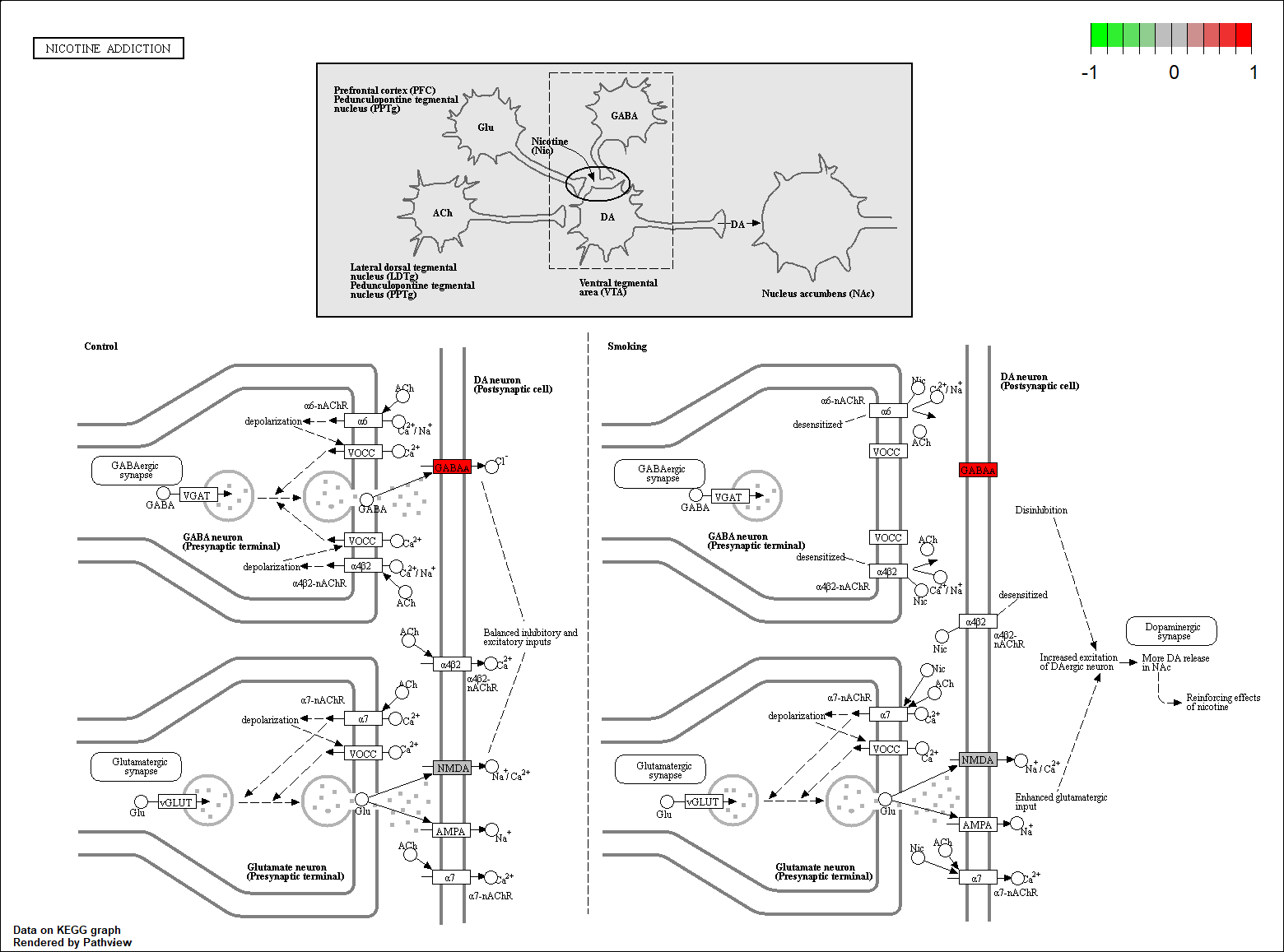
|
|
CHRNA6 |
hsa04080 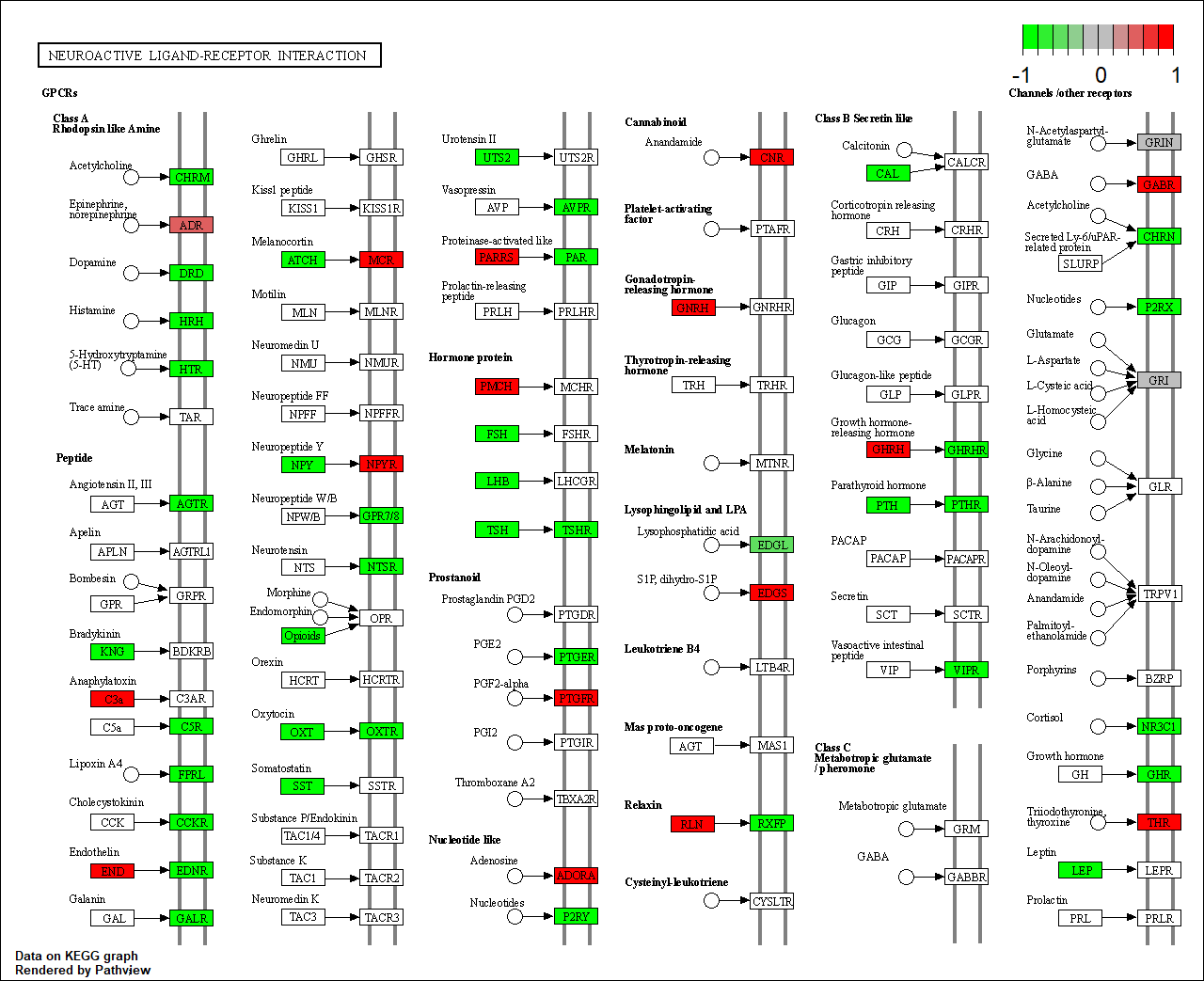
|
|
CHRNA7 
|
hsa04020 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| ZNF271P | 10.4729 | 10.6231 | 10.2614 | 6.9188 | 6.3264 |
| GINS4 | 10.2987 | 9.8827 | 9.6049 | 6.0181 | 6.1993 |
| NTAN1 | 12.5861 | 12.6886 | 11.8174 | 8.3703 | 8.4309 |
| USB1 | 9.692 | 10.6446 | 10.2897 | 6.3171 | 5.9321 |
| GSDMB | 11.7465 | 11.6297 | 12.0615 | 6.4061 | 6.0004 |
| ARHGEF28 | 10.2824 | 10.5088 | 9.6995 | 6.0031 | 6.1668 |
| ANGPTL4 | 12.5349 | 12.2657 | 11.8443 | 8.2175 | 8.357 |
| PYCARD | 13.0869 | 12.1473 | 11.9934 | 4.9375 | 5.0715 |
| APP | 13.8757 | 13.4845 | 13.1677 | 7.3934 | 6.2798 |
| SQSTM1 | 13.5502 | 13.7235 | 13.8954 | 4.3386 | 6.2798 |
| MAT2A | 13.2883 | 12.9767 | 12.4072 | 6.9369 | 8.1947 |
| CDK7 | 11.7381 | 11.5465 | 12.2007 | 5.3178 | 4.125 |
| CRK | 10.4751 | 9.7568 | 8.5992 | 3.7458 | 3.624 |
| CDC25B | 11.2927 | 11.2714 | 12.5054 | 5.4453 | 4.0461 |
| PLK1 | 10.7372 | 10.234 | 11.6194 | 5.9216 | 6.0123 |
| SNX6 | 11.8308 | 11.4439 | 12.6738 | 6.1679 | 7.0108 |
| PAF1 | 12.9149 | 12.2398 | 11.479 | 7.219 | 7.0668 |
| UBE2C | 11.7381 | 12.0391 | 13.2874 | 6.6766 | 6.15 |
| MRPL12 | 15 | 15 | 15.0004 | 8.7874 | 8.6543 |
| DDIT4 | 10.7709 | 10.9184 | 11.5758 | 6.4594 | 6.8915 |
| TMEM97 | 9.918 | 9.6624 | 10.4306 | 3.2881 | 4.5101 |
| PCMT1 | 10.0867 | 10.0168 | 9.8954 | 6.3231 | 6.15 |
| MPC2 | 11.1076 | 11.0945 | 11.8767 | 5.8006 | 6.8915 |
| ATP1B1 | 13.1374 | 12.8539 | 14.0303 | 7.6081 | 7.3861 |
| ITGB1BP1 | 8.3055 | 8.099 | 9.538 | 3.7458 | 3.8303 |
| PPP2R5E | 11.4698 | 11.0386 | 11.9263 | 3.3836 | 3.8303 |
| ENOPH1 | 11.0784 | 11.2114 | 10.2098 | 4.7427 | 5.3285 |
| NOSIP | 9.6716 | 9.8971 | 10.8543 | 5.6307 | 5.3285 |
| OXA1L | 12.5306 | 12.7953 | 13.1817 | 5.9216 | 6.0123 |
| HMGCS1 | 12.3233 | 12.0231 | 12.4621 | 4.7427 | 6.3541 |
| GTPBP8 | 11.9912 | 11.816 | 10.9305 | 5.9216 | 6.4664 |
| C2CD5 | 10.9659 | 10.7996 | 11.6029 | 6.3833 | 7.1282 |
| MRPS16 | 12.431 | 11.7714 | 13.3488 | 5.7168 | 5.8714 |
| SPR | 10.5865 | 11.0266 | 10.6558 | 4.9522 | 6.0879 |
| CCNB2 | 10.9365 | 10.3545 | 11.406 | 5.6307 | 5.8714 |
| DNM1L | 11.2236 | 10.853 | 10.0431 | 3.7458 | 5.3285 |
| COG7 | 10.7284 | 10.576 | 10.4002 | 6.5274 | 6.0879 |
| PLEKHM1 | 13.6874 | 13.3608 | 12.472 | 5.4453 | 7.1282 |
| PARK7 | 10.9086 | 11.8498 | 11.3607 | 6.0272 | 4.8329 |
| HSP90AB1 | 12.0005 | 12.3071 | 12.1127 | 7.6192 | 6.9657 |
| COX4I1 | 12.876 | 13.7187 | 14.4388 | 7.9989 | 6.6219 |
| PFN1 | 14.9418 | 15 | 15 | 11 | 11.4279 |
| HSPA9 | 14.259 | 13.9767 | 13.3583 | 8.2352 | 7.4061 |
| RAN | 14.8271 | 15 | 15 | 10.2347 | 11.1088 |
| IST1 | 10.8455 | 11.009 | 11.7032 | 6.314 | 7.1401 |
| S100A10 | 11.794 | 11.7662 | 10.7601 | 6.5444 | 6.7804 |
| DNAJA1 | 9.9859 | 10.0876 | 10.9313 | 5.2922 | 6.1655 |
| AHCY | 12.3882 | 13.6435 | 12.7494 | 7.856 | 7.8257 |
| CCT3 | 11.3005 | 12.042 | 12.3554 | 6.2816 | 5.8382 |
| SDCBP | 10.63 | 11.4219 | 11.6902 | 3.0527 | 4.593 |
| SEPHS2 | 10.4886 | 10.9237 | 10.6382 | 5.617 | 6.3509 |
| PPIB | 14.4968 | 15 | 14.155 | 7.3217 | 6.7558 |
| TAX1BP1 | 10.6474 | 10.484 | 10.8294 | 6.6639 | 6.2179 |
| NDUFB8 | 13.4012 | 13.8685 | 14.2633 | 7.9944 | 8.0108 |
| ANXA4 | 15 | 14.534 | 14.7431 | 7.4171 | 7.0972 |
| ZNHIT1 | 10.5586 | 11.4513 | 11.7289 | 5.451 | 5.6556 |
| NME1 | 15 | 15 | 15 | 9.4326 | 8.6632 |
| COPS5 | 13.1207 | 12.8251 | 12.8412 | 8.4162 | 8.0196 |
| ACSL3 | 10.7772 | 11.0554 | 9.9156 | 5.1 | 4.7771 |
| COX6C | 15 | 15 | 14.0373 | 10.2063 | 10.6324 |
| PON2 | 10.1481 | 10.7219 | 9.103 | 3.9161 | 2.3985 |
| ALDH3A2 | 12.9188 | 13.6939 | 14.0451 | 8.0392 | 7.0224 |
| NAE1 | 12.5564 | 12.5089 | 11.7482 | 5.6035 | 6.8557 |
| TK1 | 13.8663 | 13.0215 | 13.0741 | 8.7158 | 7.2201 |
| EPS8 | 12.8866 | 10.5958 | 11.9547 | 5.4913 | 4.9752 |
| SNRPD1 | 12.6502 | 13.1941 | 13.1256 | 7.8918 | 7.6604 |
| LPP | 12.2806 | 12.4526 | 11.4589 | 5.1617 | 4.4526 |
| RNASEH2A | 11.3051 | 10.9263 | 10.8123 | 6.5351 | 6.9209 |
| NAA10 | 11.3905 | 10.9418 | 11.3129 | 5.7885 | 6.7849 |
| CAV1 | 11.5021 | 12.122 | 11.2962 | 6.693 | 5.5426 |
| WTAP | 10.5781 | 10.0151 | 10.6231 | 4.3979 | 5.6089 |
| MRPL40 | 11.7512 | 11.9061 | 11.4007 | 7.6962 | 8.1035 |
| FAM50A | 10.3637 | 10.3655 | 11.9377 | 4.8797 | 5.6417 |
| SNRPE | 14.5754 | 14.3073 | 14.1314 | 9.948 | 9.0222 |
| DYNC1LI2 | 12.2528 | 12.5935 | 11.7458 | 6.9858 | 6.1952 |
| ZWINT | 14.2745 | 14.6288 | 14.1529 | 7.8662 | 8.6901 |
| CKS2 | 10.7805 | 11.1377 | 12.7654 | 5.3292 | 4.8411 |
| LGALS4 | 15 | 15 | 14.7201 | 6.3283 | 2.2017 |
| MRPS12 | 15 | 14.8659 | 15 | 9.335 | 8.8714 |
| POP5 | 10.8959 | 11.281 | 11.2148 | 6.5083 | 7.3613 |
| HSPE1 | 13.4534 | 13.7875 | 12.4633 | 5.5497 | 6.4838 |
| SNRPA1 | 11.8076 | 11.2734 | 11.5886 | 5.2648 | 6.3361 |
| OPHN1 | 13.9981 | 13.4802 | 13.2957 | 5.7247 | 6.5451 |
| AKR1B10 | 14.3512 | 12.4502 | 15 | 3.858 | 0.8259 |
| ABCC3 | 9.8782 | 11.0128 | 9.948 | 3.011 | 4.053 |
| YBX1 | 10.6646 | 10.7201 | 11.6008 | 6.7584 | 6.5433 |
| HMGN2 | 12.8004 | 12.3773 | 12.5737 | 8.3149 | 8.112 |
| RBM39 | 10.7484 | 11.5342 | 10.6975 | 5.0312 | 5.8136 |
| C1S | 6.9682 | 6.236 | 7.0957 | 2.051 | 1.211 |
| ATIC | 10.7577 | 10.2614 | 10.376 | 6.1409 | 5.9928 |
| PSMA6 | 13.776 | 12.8347 | 12.0989 | 7.1877 | 7.0095 |
| PSMB6 | 14.2141 | 14.7201 | 15 | 9.4123 | 9.6126 |
| TXN | 15 | 13.5286 | 14.2171 | 8.2584 | 6.6287 |
| C1QBP | 14.0516 | 15 | 14.2076 | 7.0456 | 6.4162 |
| ID1 | 8.7405 | 8.8179 | 7.7321 | 2.0801 | 2.4504 |
| DUT | 11.3761 | 12.0909 | 11.4764 | 6.0906 | 5.5565 |
| RSRP1 | 9.2399 | 9.2123 | 9.9044 | 4.0549 | 3.6496 |
| OSGEP | 11.8345 | 11.5199 | 11.9859 | 7.6394 | 7.6077 |
| APEX1 | 13.9018 | 13.9798 | 15 | 6.8486 | 6.8114 |
| TPX2 | 10.2913 | 10.1046 | 10.4931 | 5.509 | 6.1406 |
| MAGOH | 11.4646 | 12.0178 | 11.784 | 4.5689 | 6.1219 |
| ADSL | 13.8382 | 14.2673 | 13.3483 | 8.851 | 8.8862 |
| NUDC | 11.7742 | 11.818 | 11.9759 | 6.0074 | 7.1456 |
| ASPH | 13.82 | 15 | 13.6981 | 7.6019 | 7.2332 |
| LRPPRC | 11.6703 | 11.6524 | 12.3266 | 7.9877 | 7.3541 |
| KPNA2 | 14.5979 | 15 | 14.0698 | 7.0442 | 6.7537 |
| HSPA5 | 13.11 | 12.8175 | 12.3233 | 7.281 | 7.9332 |
| HSP90AA1 | 13.0777 | 13.4404 | 13.8227 | 4.2869 | 4.8886 |
| RSL1D1 | 10.8178 | 10.4297 | 10.2486 | 4.984 | 6.1962 |
| CBX5 | 10.9744 | 10.4615 | 10.4899 | 6.388 | 6.0102 |
| FKBP9 | 11.5675 | 12.1977 | 11.573 | 6.6343 | 6.2697 |
| TSR3 | 11.6048 | 10.4878 | 10.9447 | 6.3932 | 6.4438 |
| NPTX2 | 15 | 15 | 15 | 11.1875 | 10.6396 |
| COX5B | 13.4737 | 13.475 | 14.0156 | 8.8754 | 8.9955 |
| NOP2 | 10.5277 | 10.5824 | 11.2341 | 6.2601 | 6.1342 |
| PQBP1 | 12.5478 | 13.1235 | 12.6866 | 7.8694 | 6.5728 |
| STOML2 | 15 | 14.7578 | 15 | 9.3351 | 10.3946 |
| TUBA3D | 10.1897 | 11.0189 | 11.0568 | 5.9288 | 6.408 |
| NUCKS1 | 12.3986 | 11.7728 | 11.6302 | 6.6525 | 6.6688 |
| MRPL18 | 11.1112 | 10.995 | 11.4178 | 6.5065 | 6.4562 |
| MRPS35 | 12.6326 | 12.1738 | 12.1399 | 6.3341 | 7.2093 |
| FKBP3 | 12.5089 | 11.618 | 11.9461 | 6.6089 | 6.5438 |
| DCTPP1 | 14.3354 | 13.6049 | 13.1753 | 8.1778 | 7.9773 |
| HMOX2 | 10.6957 | 11.4899 | 10.5564 | 6.2161 | 6.128 |
| RBM22 | 10.0965 | 10.3957 | 10.2569 | 6.7621 | 6.4732 |
| MRPL22 | 13.7561 | 13.5012 | 12.8974 | 6.468 | 7.5007 |
| ZWILCH | 10.3217 | 10.1998 | 9.4094 | 5.7192 | 4.8477 |
| CMC2 | 10.7898 | 10.0529 | 10.3844 | 5.824 | 6.4855 |
| YARS2 | 12.7686 | 12.1992 | 11.7776 | 7.0601 | 7.6135 |
| CHORDC1 | 9.8702 | 10.118 | 9.3945 | 5.2908 | 3.564 |
| TSPAN15 | 10.4798 | 10.6464 | 10.9802 | 7.1739 | 7.2095 |
| PTRH2 | 10.5686 | 11.6379 | 10.6628 | 6.6384 | 6.0927 |
| INTS8 | 9.2004 | 9.5972 | 8.6165 | 5.0733 | 4.0922 |
| SYNJ2BP | 10.7284 | 10.6752 | 10.15 | 5.4 | 5.1233 |
| OLA1 | 10.8854 | 11.3866 | 10.7204 | 6.7407 | 6.7955 |
| EMC6 | 12.3206 | 12.7302 | 12.8164 | 7.2582 | 8.6188 |
| METTL5 | 10.6257 | 10.9737 | 10.379 | 4.6868 | 6.0409 |
| BABAM1 | 11.1285 | 11.648 | 12.2972 | 7.093 | 6.4532 |
| KAT8 | 9.4286 | 9.6357 | 9.9916 | 6.0845 | 6.0555 |
| TRMT5 | 12.9835 | 13.5069 | 13.0408 | 8.9389 | 9.0602 |
| OTUB1 | 10.9507 | 11.838 | 12.2688 | 7.0956 | 6.5722 |
| LTB | 9.7938 | 11.0151 | 9.4973 | 3.6032 | 2.2486 |
PTPN11 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: PTPN11
[1] [PTPN11 and the deafness].
[2] Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment.
[3] The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia.
[8] Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling.
[9] PTPN11 hypomethylation is associated with gastric cancer progression.
[10] A PTPN11 mutation in a woman with Noonan syndrome and protein-losing enteropathy.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa04722