BHLHE40 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1654
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: imatinib
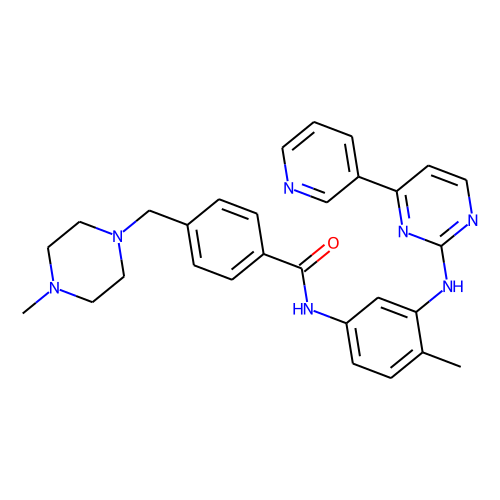
| ID | BRD-K92723993 |
| name | imatinib |
| type | Small molecule |
| molecular formula | C29H31N7O |
| molecular weight | 493.6 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | CN1CCN(Cc2ccc(cc2)C(=O)Nc2ccc(C)c(Nc3nccc(n3)-c3cccnc3)c2)CC1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: A549
Cell lines used for experiments
TRANSCRIPTION FACTOR: BHLHE40
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
ABL1 
|
hsa05200 
|
|
CSF1R 
|
hsa04015 
|
|
DDR1 
|
|
|
KIT 
|
hsa04916 
|
|
NTRK1 
|
hsa04014 
|
|
PDGFRA 
|
|
|
PDGFRB 
|
hsa04510 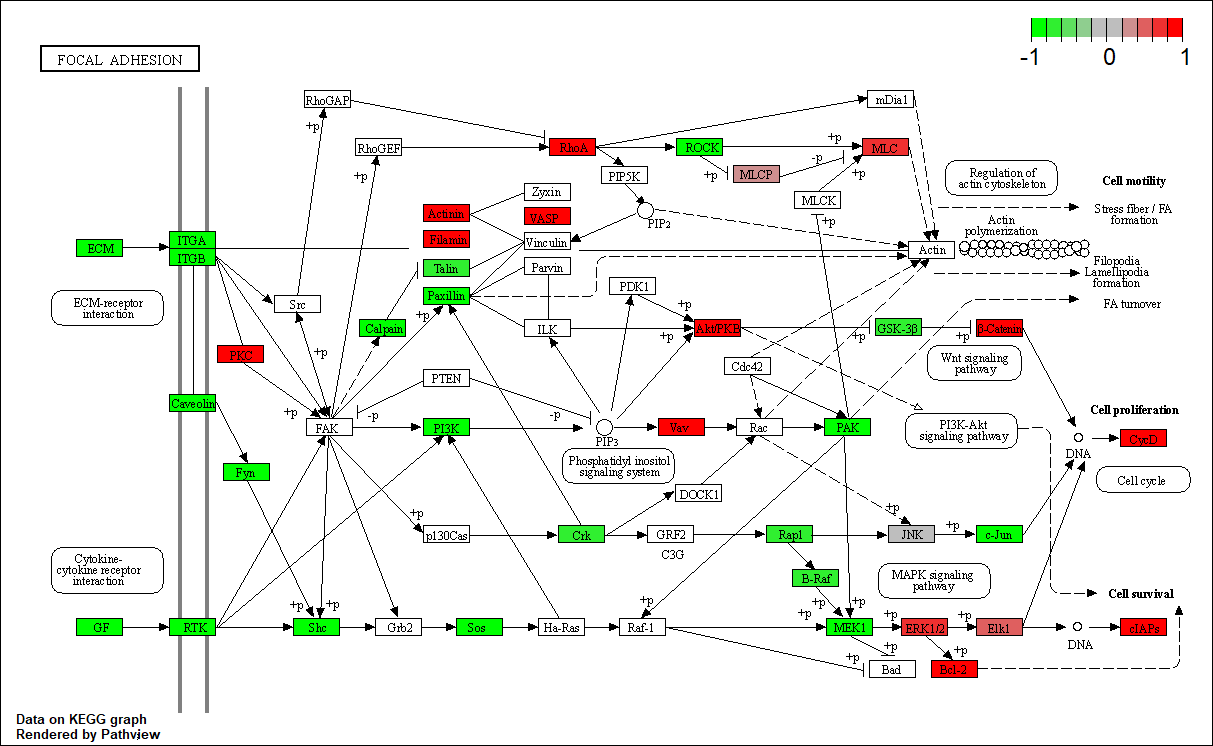
|
|
RET 
|
hsa05223 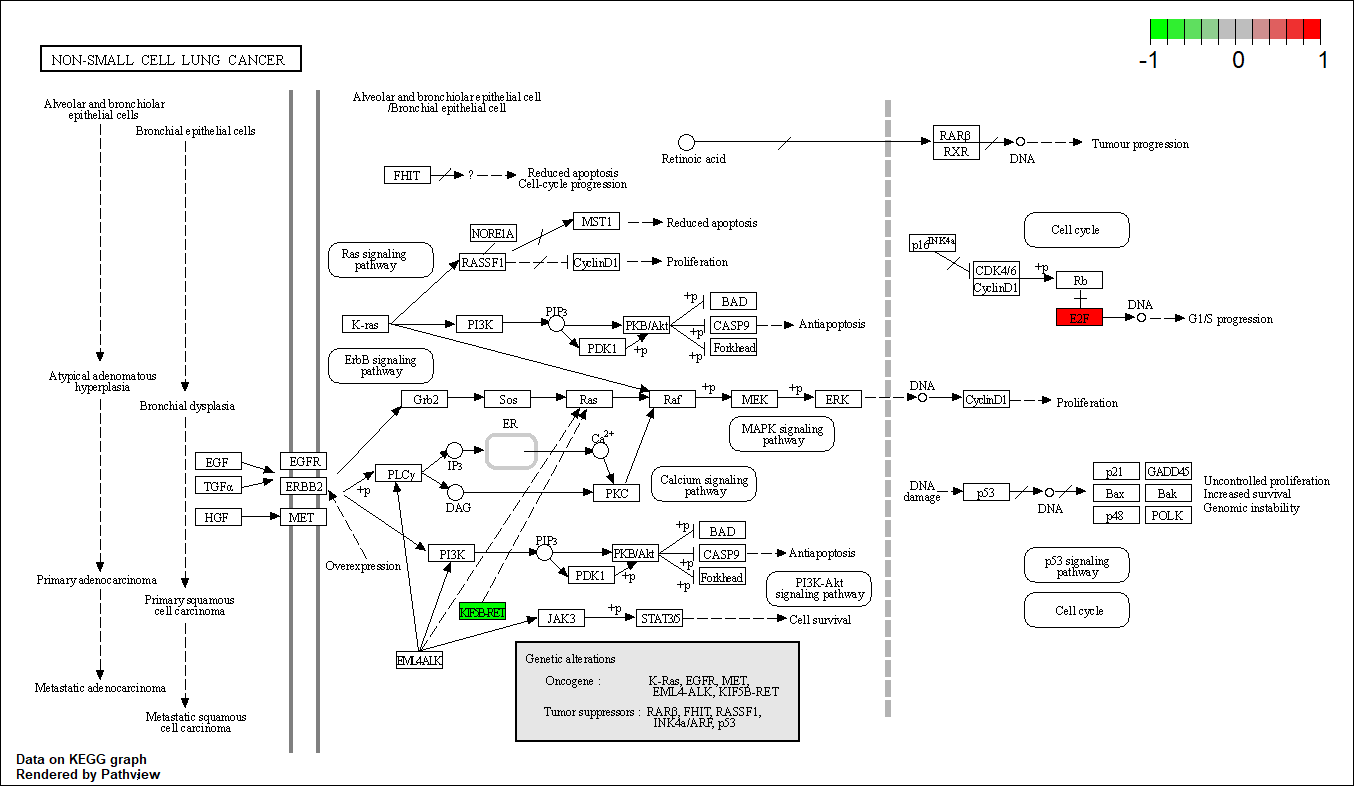
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition |
|---|---|---|---|---|
| ZFHX3 | 7.4738 | 7.7471 | 7.8971 | 9.2134 |
| LGALS2 | 7.3394 | 7.4429 | 7.2361 | 8.6 |
| FAM107A | 6.0027 | 5.0537 | 4.8616 | 7.3231 |
| SPRY1 | 10.7729 | 11.0513 | 10.5766 | 12.5688 |
| KDM6B | 9.0743 | 9.1166 | 9.1889 | 10.2115 |
| CCDC88A | 8.3647 | 8.7311 | 8.7151 | 9.8121 |
| IL17RC | 6.8151 | 6.8003 | 6.9753 | 7.9814 |
| BCAM | 10.7216 | 10.7552 | 10.8687 | 12.2647 |
| SERPINE1 | 6.3872 | 6.4235 | 7.0019 | 8.0478 |
| EIF4G1 | 8.7429 | 9.2103 | 9.4151 | 13.978 |
| SIRT3 | 6.1177 | 6.1637 | 5.7527 | 7.5626 |
| CXCL2 | 4.5613 | 4.1122 | 4.6157 | 5.7908 |
| PAK4 | 8.0998 | 7.6446 | 7.7066 | 8.9878 |
| BHLHE40 | 3.8189 | 3.0833 | 3.5752 | 5.5898 |
| TBPL1 | 6.9302 | 7.023 | 6.4757 | 8.1985 |
| PLA2G15 | 4.7268 | 4.6553 | 4.4681 | 6.0738 |
| ARHGEF2 | 5.7337 | 5.7039 | 5.5954 | 6.9691 |
| CPNE3 | 13.3556 | 13.0089 | 13.004 | 14.4123 |
| NOLC1 | 9.518 | 9.3832 | 9.4329 | 10.5907 |
| POLD4 | 10.2531 | 10.3207 | 9.6375 | 11.5819 |
| RGS2 | 8.4524 | 8.1663 | 7.9147 | 9.6093 |
| TSEN2 | 8.4202 | 8.0071 | 7.8053 | 9.5026 |
| SLC35F2 | 6.8821 | 5.8269 | 6.7084 | 8.8356 |
| NUDT9 | 10.6082 | 10.3038 | 10.7103 | 12.0587 |
| TLK2 | 10.4245 | 10.8509 | 10.3846 | 11.8615 |
| PNKP | 6.8458 | 6.7183 | 7.109 | 8.7428 |
| SLC35A3 | 5.8492 | 5.9534 | 5.259 | 7.4577 |
| MTA1 | 8.7163 | 7.9788 | 7.8943 | 10.8127 |
| P4HTM | 6.8458 | 6.1284 | 6.8042 | 8.0983 |
| ID2 | 7.119 | 6.019 | 6.1501 | 8.4283 |
| INTS3 | 8.9751 | 8.8657 | 8.327 | 11.2887 |
| FASTKD5 | 8.4202 | 8.107 | 8.6276 | 11.0724 |
| GYS1 | 8.2906 | 7.2854 | 7.416 | 9.7749 |
| GDI2 | 7.966 | 7.4451 | 7.4754 | 9.0145 |
| ACTN4 | 13.1142 | 12.1588 | 13.057 | 14.4546 |
| CTSB | 7.0626 | 7.9356 | 7.2168 | 9.0418 |
| ABLIM1 | 10.7827 | 10.6252 | 11.2074 | 13.2366 |
| ARHGDIA | 9.3067 | 9.5237 | 9.6593 | 10.6881 |
| AMD1 | 5.445 | 4.8826 | 5.7335 | 6.9232 |
| HNRNPAB | 12.6241 | 13.2786 | 12.9775 | 14.2326 |
| PACSIN2 | 11.4812 | 11.9745 | 12.3587 | 13.6276 |
| LRRFIP1 | 7.4712 | 7.0081 | 6.7732 | 9.8864 |
| ALCAM | 2.5533 | 2.0925 | 3.6934 | 6.0673 |
| CIB1 | 8.9474 | 8.328 | 8.6679 | 9.9032 |
| CTR9 | 5.7292 | 6.1213 | 5.7066 | 6.9969 |
| PLIN3 | 7.4729 | 7.1782 | 7.2349 | 9.0348 |
| FADS2 | 10.7201 | 10.3353 | 11.2606 | 13.0586 |
| RAB13 | 12.0904 | 11.6123 | 12.5611 | 13.9876 |
| KLF10 | 5.8049 | 5.8425 | 5.7327 | 7.0497 |
| LSM4 | 8.5581 | 8.865 | 8.516 | 9.8339 |
| GPRC5A | 6.7027 | 5.8402 | 7.2638 | 9.0775 |
| CFDP1 | 10.4927 | 10.6497 | 10.3167 | 12.2163 |
| CNOT3 | 9.2866 | 9.1319 | 9.5849 | 10.672 |
| CAPN7 | 5.3814 | 5.2149 | 5.1271 | 6.3176 |
| PDLIM7 | 11.2766 | 11.2759 | 11.4538 | 12.48 |
| PMVK | 9.4089 | 8.6705 | 8.8888 | 10.8475 |
| ALG8 | 7.2906 | 7.2383 | 6.9641 | 8.4506 |
| TERF2 | 7.1684 | 7.9837 | 7.247 | 9.6639 |
| RARA | 5.8448 | 5.3048 | 5.8572 | 7.2533 |
| GCLM | 8.8268 | 8.6923 | 9.1362 | 10.3266 |
| PHACTR2 | 4.8811 | 5.7136 | 5.7424 | 7.3315 |
| RBM14 | 7.9347 | 7.375 | 7.9581 | 9.375 |
| FGFR3 | 3.4186 | 4.076 | 3.7934 | 5.727 |
| PC | 6.3239 | 6.6303 | 6.3605 | 7.5254 |
| HPS5 | 6.3172 | 6.7744 | 6.4462 | 8.0087 |
| SLC7A7 | 11.3429 | 10.9775 | 11.0175 | 12.382 |
| BCL3 | 6.4348 | 5.3717 | 6.3115 | 8.8064 |
| SLC26A2 | 0 | 0 | 0 | 2.2825 |
| WASL | 10.1592 | 10.6532 | 9.7587 | 11.7059 |
| SOCS3 | 4.9467 | 4.8165 | 5.011 | 6.4559 |
| SIX2 | 7.9342 | 7.0532 | 7.2492 | 9.5828 |
| NFIC | 6.6678 | 6.7281 | 6.3818 | 8.1576 |
| NCOR2 | 8.173 | 8.3757 | 8.0254 | 10.3167 |
| ATP6AP1 | 6.1201 | 6.4806 | 6.6189 | 7.5702 |
| SF1 | 9.9202 | 9.7858 | 10.1047 | 11.3256 |
| NAPA | 8.9144 | 8.1978 | 8.3314 | 9.8541 |
| POR | 8.6693 | 8.5991 | 8.3842 | 10.0796 |
| NR2F2 | 4.5708 | 5.2255 | 4.6781 | 6.6431 |
| ITPA | 10.8067 | 10.6974 | 10.9047 | 11.7927 |
| SEC23IP | 6.5471 | 6.57 | 6.6265 | 7.9461 |
| IRS2 | 5.8466 | 5.5574 | 6.4183 | 7.4492 |
| PIM1 | 9.8414 | 10.0219 | 9.9478 | 11.3934 |
| CLN3 | 8.757 | 8.2029 | 8.2238 | 9.6498 |
| PPAT | 5.2049 | 5.5603 | 5.4899 | 6.6179 |
| HTATIP2 | 7.0197 | 7.9282 | 7.8767 | 9.5954 |
| DHTKD1 | 6.6018 | 6.5399 | 5.837 | 7.9377 |
| MBP | 8.9017 | 8.5949 | 8.5026 | 10.7325 |
| SLC19A1 | 6.4523 | 6.4617 | 6.6556 | 7.763 |
| FASN | 10.3241 | 10.0026 | 11.1025 | 12.8449 |
| PBXIP1 | 8.1921 | 8.4482 | 8.4193 | 9.4663 |
| NEK9 | 6.59 | 6.6484 | 6.4876 | 7.9235 |
| SETD3 | 6.5443 | 6.9602 | 6.3188 | 8.0205 |
| HPCAL1 | 11.5149 | 11.3136 | 11.2667 | 12.5599 |
| EHBP1 | 5.8082 | 5.0699 | 5.8463 | 7.3144 |
| LARP4 | 6.4895 | 7.1301 | 6.6827 | 8.1062 |
| SIK3 | 3.7576 | 3.4539 | 3.484 | 4.6165 |
| MCAT | 8.6424 | 8.7105 | 8.6456 | 9.9104 |
| KLC1 | 7.247 | 7.3195 | 7.4994 | 8.5052 |
| MLX | 6.9527 | 6.6393 | 6.781 | 7.8342 |
| PCGF2 | 6.5293 | 5.5707 | 6.3966 | 8.424 |
| TADA3 | 7.7635 | 8.2721 | 7.9062 | 9.2103 |
| MT1M | 6.3429 | 6.9173 | 6.8847 | 8.0799 |
| TNS3 | 8.0653 | 6.9215 | 8.7458 | 10.6639 |
| LIMA1 | 6.9722 | 7.1827 | 7.331 | 8.7657 |
| PDXK | 6.4026 | 6.0314 | 5.9456 | 7.2885 |
| RBM47 | 4.3532 | 3.7576 | 3.5773 | 6.8995 |
| MRPS34 | 10.606 | 9.9633 | 10.3563 | 11.7596 |
| SCAMP2 | 5.1249 | 4.8987 | 5.1817 | 6.4601 |
| GALNT7 | 7.5892 | 8.9819 | 8.5252 | 10.4797 |
| JADE1 | 5.0353 | 4.3142 | 4.9717 | 6.2997 |
| C1orf115 | 6.1778 | 6.8599 | 6.4614 | 8.7174 |
| TRPM4 | 4.2604 | 3.1189 | 3.317 | 5.8589 |
| TMEM164 | 5.7085 | 6.1009 | 5.4126 | 7.3824 |
| EHBP1L1 | 5.8251 | 6.2766 | 6.004 | 7.8116 |
| ABHD2 | 6.2112 | 6.6587 | 7.1774 | 8.9984 |
| THOC2 | 6.4924 | 5.8184 | 5.705 | 7.5481 |
| EXOC1 | 7.0245 | 6.4462 | 6.7881 | 8.3264 |
| SLC35E1 | 6.6311 | 7.0058 | 7.0113 | 8.4315 |
| SFTPC | 4.469 | 4.6681 | 3.882 | 5.988 |
| GMFG | 6.627 | 6.5843 | 6.2281 | 7.6639 |
| AHDC1 | 9.5832 | 9.2719 | 9.4762 | 10.6001 |
| PTP4A3 | 9.7898 | 10.6943 | 10.3693 | 12.0023 |
BHLHE40 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: BHLHE40
[1] Transcription Factor Bhlhe40 in Immunity and Autoimmunity.
[4] Bhlhe40 is an essential repressor of IL-10 during Mycobacterium tuberculosis infection.
[9] TFEB Transcriptional Responses Reveal Negative Feedback by BHLHE40 and BHLHE41.
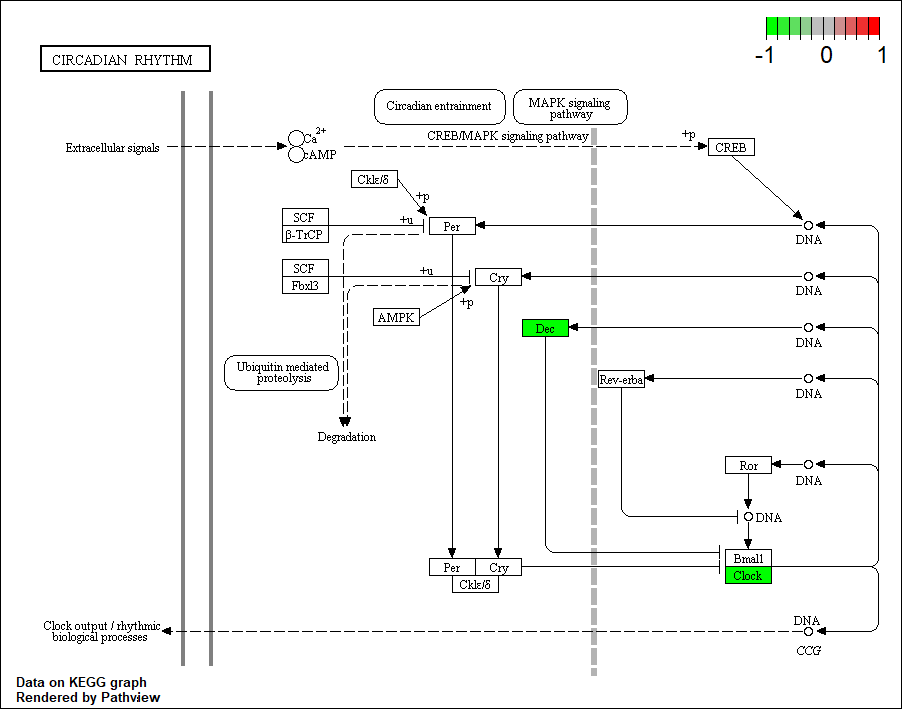
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa04710