KLF4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1716
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: BRD-A84102390
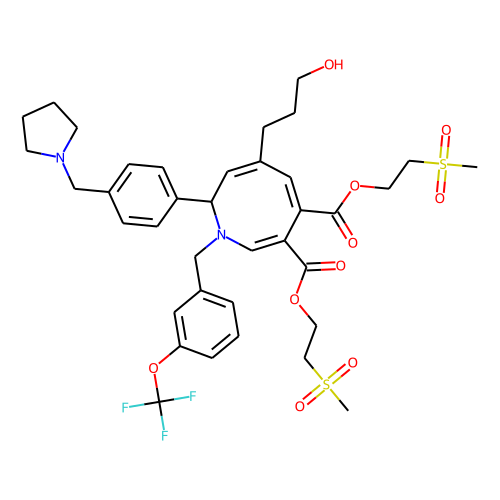
| ID | BRD-A84102390 |
| name | BRD-A84102390 |
| type | Small molecule |
| molecular formula | NA |
| molecular weight | NA |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | CS(=O)(=O)CCOC(=O)C1=CN(Cc2cccc(OC(F)(F)F)c2)C(C=C(CCCO)C=C1C(=O)OCCS(C)(=O)=O)c1ccc(CN2CCCC2)cc1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: KLF4
Transcription regulatory factors corresponding to this network
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|---|
| ATF1 | 7.6127 | 8.275 | 8.5414 | 5.0419 | 4.7798 | 4.6768 | 5.3075 | 5.3232 |
| CEBPA | 6.9966 | 6.3484 | 5.6684 | 2.9781 | 3.3658 | 2.5262 | 3.131 | 3.2205 |
| PCNA | 14.4011 | 14.3895 | 14.0599 | 7.9914 | 8.5499 | 7.9301 | 8.1182 | 10.4845 |
| CDC20 | 13.8848 | 13.9308 | 13.8223 | 9.9291 | 11.3667 | 10.2592 | 10.3187 | 11.2449 |
| CDC25B | 12.5588 | 11.7754 | 11.1411 | 7.573 | 8.6702 | 7.1074 | 8.153 | 7.4013 |
| SPDEF | 12.8931 | 12.6931 | 12.2454 | 10.1269 | 10.4575 | 10.2258 | 10.3881 | 10.5163 |
| GABPB1 | 9.6294 | 9.5543 | 9.7403 | 5.1731 | 5.4416 | 5.3182 | 4.6337 | 5.3033 |
| SOX4 | 9.3283 | 9.0105 | 8.1053 | 4.6516 | 4.8285 | 4.8752 | 5.2748 | 5.136 |
| UBE2C | 12.4569 | 12.6742 | 12.8998 | 7.3541 | 8.747 | 7.5427 | 9.3917 | 8.3636 |
| TXLNA | 9.7277 | 10.2764 | 9.9282 | 5.4733 | 6.3997 | 5.6446 | 6.9394 | 5.5975 |
| DNAJB1 | 9.9955 | 10.3472 | 10.5258 | 6.4096 | 5.2396 | 5.5774 | 5.7406 | 5.4754 |
| CCNA2 | 9.9359 | 9.9197 | 9.795 | 4.7756 | 5.2774 | 4.4181 | 5.3075 | 4.8383 |
| VAPB | 9.1483 | 9.0949 | 9.3553 | 6.876 | 7.5666 | 6.5602 | 7.1205 | 6.7954 |
| ATMIN | 8.6324 | 8.5089 | 6.9896 | 4.4001 | 4.3162 | 4.2106 | 4.1045 | 3.9223 |
| GLOD4 | 10.1947 | 10.5125 | 10.1215 | 6.4096 | 7.0992 | 6.6079 | 6.5844 | 6.52 |
| TMEM97 | 9.6167 | 9.5103 | 9.4039 | 6.9271 | 6.6349 | 6.0796 | 6.5208 | 5.7649 |
| B4GAT1 | 8.896 | 8.7556 | 8.7728 | 5.3805 | 5.2053 | 5.1552 | 5.0536 | 6.2135 |
| RAD9A | 7.4524 | 6.9349 | 7.0357 | 3.7432 | 3.6793 | 3.4072 | 4.2211 | 4.1045 |
| RAE1 | 11.4059 | 11.8962 | 12.104 | 8.5124 | 9.3411 | 8.9393 | 9.5032 | 8.7594 |
| BCL7B | 6.4826 | 6.167 | 6.3667 | 4.2525 | 4.523 | 3.9174 | 4.3628 | 4.1676 |
| PLA2G15 | 6.9253 | 6.3804 | 6.1721 | 4.544 | 4.4218 | 4.8116 | 4.3628 | 4.2774 |
| PRSS23 | 11.3418 | 11.5896 | 11.0659 | 8.6507 | 9.0375 | 8.3464 | 8.738 | 8.0087 |
| RRP1B | 7.9552 | 7.8372 | 7.7444 | 5.4103 | 5.0573 | 5.8551 | 4.6337 | 5.4917 |
| BLVRA | 10.1143 | 9.2717 | 9.7849 | 6.7243 | 7.6077 | 6.6513 | 7.309 | 6.82 |
| NET1 | 9.72 | 10.0442 | 9.8052 | 6.5213 | 6.7613 | 6.7273 | 6.1714 | 6.4852 |
| NCOA3 | 13.4753 | 12.9899 | 12.8324 | 10.6518 | 10.4478 | 10.7756 | 10.9349 | 10.8771 |
| VGLL4 | 9.2744 | 8.934 | 8.7296 | 6.4879 | 6.8313 | 6.4887 | 6.3576 | 7.0324 |
| CANT1 | 13.1 | 12.6381 | 12.417 | 10.7327 | 11 | 10.8843 | 10.8321 | 10.8795 |
| TM9SF3 | 9.7845 | 8.534 | 9.1302 | 6.1415 | 6.106 | 6.0099 | 6.8142 | 5.6474 |
| PARP2 | 9.4143 | 9.2608 | 9.1021 | 4.8907 | 4.8998 | 5.2706 | 5.4026 | 4.5735 |
| RNPS1 | 15 | 13.6293 | 14.3757 | 10.8662 | 11.3176 | 11.1552 | 11.4911 | 11.8449 |
| PWP1 | 11.2162 | 11.1038 | 10.8297 | 9.217 | 8.5054 | 8.2777 | 9.0785 | 8.7463 |
| IMPA2 | 7.8771 | 8.6794 | 8.164 | 4.8454 | 4.086 | 4.5858 | 5.6242 | 4.0078 |
| DLGAP5 | 9.3999 | 9.888 | 9.6901 | 6.0889 | 6.8009 | 6.9634 | 6.9721 | 6.9958 |
| GDI2 | 10.8192 | 10.3777 | 10.7598 | 7.5615 | 7.2785 | 7.2053 | 7.1862 | 8.5676 |
| NPC2 | 7.3831 | 7.2639 | 6.9468 | 4.5277 | 5.3315 | 4.6172 | 4.3529 | 5.0433 |
| RAN | 14.9921 | 14.8313 | 15 | 13.1351 | 13.0665 | 13.0133 | 12.9851 | 13.2383 |
| ARL6IP5 | 8.2535 | 7.9465 | 7.5535 | 4.5683 | 4.898 | 5.3884 | 4.8661 | 4.2904 |
| AMD1 | 8.1324 | 7.5814 | 7.2471 | 3.8066 | 2.2796 | 4.4729 | 3.929 | 3.7344 |
| RAF1 | 8.4825 | 8.3773 | 8.6041 | 5.6865 | 6.4692 | 5.4635 | 5.6365 | 5.899 |
| ANP32B | 11.9072 | 11.8438 | 11.6021 | 7.7333 | 9.0795 | 8.452 | 8.4058 | 8.9542 |
| PPP1R12A | 8.6866 | 8.6218 | 8.0269 | 5.7757 | 5.5895 | 5.4154 | 5.9263 | 5.7617 |
| CNIH1 | 11.7388 | 12.0306 | 11.8384 | 9.6528 | 9.9511 | 9.8497 | 9.5694 | 10.2915 |
| PRKAG1 | 8.4843 | 8.4974 | 8.1723 | 6.4784 | 6.3707 | 5.8108 | 5.7674 | 6.4072 |
| CTR9 | 6.7811 | 7.5133 | 6.9075 | 4.2211 | 4.6979 | 4.1989 | 4.3873 | 4.5579 |
| PRPF3 | 8.5052 | 8.9591 | 8.3533 | 6.1109 | 6.0552 | 5.9875 | 6.5928 | 6.3046 |
| CYP1B1 | 10.7003 | 10.4046 | 8.5154 | 2.788 | 3.793 | 3.1363 | 3.2815 | 4.5753 |
| AFG3L2 | 9.3145 | 9.2297 | 8.4992 | 5.9906 | 6.625 | 6.2944 | 6.4766 | 5.7334 |
| SHOC2 | 6.2243 | 5.633 | 5.7499 | 2.7985 | 3.6572 | 3.4009 | 3.6909 | 3.1272 |
| SLPI | 4.4903 | 4.4293 | 5.3608 | 1.0866 | 1.0866 | 1.4832 | 1.5161 | 0 |
| BCAS2 | 11.6643 | 11.945 | 11.496 | 9.0793 | 8.9991 | 9.5618 | 8.8431 | 9.6691 |
| CUL2 | 10.1136 | 10.3955 | 10.3128 | 7.6828 | 8.0175 | 8.136 | 7.1576 | 7.8234 |
| BMS1 | 9.9694 | 10.0223 | 9.5607 | 7.2994 | 7.5704 | 7.2953 | 6.473 | 6.7709 |
| SNRPE | 15 | 14.9648 | 15 | 12.4646 | 12.8368 | 11.687 | 12.5152 | 12.4747 |
| UPF2 | 7.141 | 7.0715 | 7.0958 | 4.3953 | 4.141 | 4.6191 | 4.4013 | 5.1168 |
| POLR2D | 8.2698 | 8.04 | 7.8633 | 6.0604 | 6.3237 | 5.8957 | 6.3054 | 5.9615 |
| ZNF263 | 8.3666 | 7.8748 | 8.042 | 6.5198 | 6.2626 | 6.4665 | 6.3659 | 6.2234 |
| IGFBP6 | 7.262 | 7.3618 | 7.3109 | 5.5116 | 5.2822 | 5.3843 | 5.4823 | 5.332 |
| VRK1 | 9.1219 | 9.405 | 8.7473 | 5.6147 | 6.1738 | 5.7319 | 5.8128 | 5.7961 |
| NR1H3 | 7.708 | 7.2428 | 7.1708 | 5.1219 | 5.3436 | 5.0724 | 4.5384 | 5.003 |
| CKS2 | 11.1666 | 11.3892 | 11.732 | 6.502 | 6.7721 | 7.3249 | 8.4869 | 7.9593 |
| PPM1D | 9.9072 | 9.6238 | 9.285 | 6.5266 | 5.5786 | 6.7444 | 6.9297 | 6.9391 |
| IDI1 | 11.7016 | 11.9525 | 11.6056 | 7.494 | 6.0192 | 7.1383 | 6.2354 | 6.2289 |
| FEN1 | 12.2346 | 13.1514 | 11.9139 | 9.4441 | 9.299 | 8.9702 | 9.4876 | 9.6005 |
| ORC2 | 7.3983 | 7.5729 | 7.3945 | 5.499 | 5.7028 | 5.528 | 5.9704 | 5.3966 |
| EXOSC9 | 7.4628 | 7.9529 | 7.0852 | 5.491 | 5.3609 | 5.2982 | 5.1754 | 5.4474 |
| PITRM1 | 8.9556 | 8.6123 | 8.4383 | 5.1431 | 5.2485 | 5.5468 | 5.7014 | 5.2273 |
| STIL | 7.6002 | 7.9879 | 7.4482 | 5.1 | 5.2436 | 5.2649 | 5.8325 | 5.7976 |
| ATXN3 | 7.2306 | 7.7048 | 6.9139 | 5.2917 | 5.3566 | 5.3386 | 5.3695 | 5.2405 |
| SNAPC1 | 7.0017 | 8.3464 | 6.7404 | 3.8832 | 4.2551 | 3.9636 | 4.1605 | 4.0195 |
| SAC3D1 | 9.7421 | 8.6739 | 9.0601 | 5.6137 | 6.1129 | 5.0374 | 6.2197 | 5.7587 |
| GEMIN4 | 9.3364 | 8.7423 | 8.8993 | 6.4887 | 6.6344 | 6.5265 | 6.7141 | 6.3659 |
| FXYD2 | 7.8798 | 7.9167 | 7.6205 | 5.0061 | 5.2517 | 4.4414 | 5.2566 | 4.1418 |
| ZC3H11A | 8.934 | 8.6779 | 8.5789 | 6.1181 | 6.5445 | 6.5932 | 7.0055 | 6.8849 |
| YAF2 | 6.987 | 6.7522 | 6.7638 | 5.111 | 5.3048 | 4.8406 | 4.7002 | 5.1046 |
| HOXC6 | 6.0221 | 4.9931 | 3.4984 | 0 | 0.1177 | 0 | 0 | 1.1569 |
| EHD1 | 8.0191 | 7.6034 | 7.2965 | 5.618 | 5.8036 | 5.5244 | 5.2303 | 5.7645 |
| STK24 | 8.0027 | 8.1198 | 7.8373 | 5.5842 | 5.1723 | 5.8345 | 5.671 | 5.5912 |
| SEPHS1 | 9.173 | 9.0538 | 9.1026 | 6.4503 | 6.8281 | 6.5282 | 6.558 | 7.2214 |
| GGA3 | 8.5177 | 8.5996 | 8.9172 | 6.2987 | 6.3493 | 6.0224 | 6.4597 | 6.4788 |
| PIGH | 8.2932 | 8.2869 | 7.887 | 6.0367 | 5.9842 | 5.8534 | 6.215 | 6.5292 |
| CDKN3 | 10.524 | 10.7973 | 10.5891 | 5.8328 | 6.6295 | 5.8284 | 6.4011 | 7.995 |
| MED6 | 8.7896 | 8.5653 | 8.7857 | 5.628 | 5.3902 | 5.624 | 5.3475 | 5.7924 |
| TUBD1 | 10.0615 | 10.3389 | 10.8076 | 7.1017 | 6.8011 | 7.5698 | 7.3987 | 7.2497 |
| EIF4A1 | 13.6434 | 14.0496 | 13.4135 | 11.655 | 11.945 | 11.73 | 11.4133 | 11.1703 |
| ARL6IP1 | 9.5845 | 9.4151 | 8.8415 | 6.1663 | 5.7267 | 6.5482 | 5.6273 | 5.1776 |
| MRFAP1L1 | 8.3188 | 8.0109 | 7.2585 | 4.0546 | 4.8405 | 4.7011 | 4.7454 | 5.3901 |
| HACD2 | 7.7868 | 8.4865 | 8.364 | 5.7781 | 5.7459 | 5.9125 | 6.0899 | 5.186 |
| NME4 | 11.4622 | 10.4186 | 10.3899 | 7.9198 | 7.3681 | 8.0342 | 7.6484 | 7.225 |
| RCHY1 | 6.5224 | 6.5717 | 6.1574 | 3.0998 | 3.5496 | 3.8281 | 3.3292 | 4.1885 |
| ZBTB1 | 7.9641 | 8.0659 | 7.8608 | 5.2485 | 5.4611 | 5.7634 | 5.2595 | 5.9418 |
| MTMR4 | 9.0179 | 8.2246 | 8.3493 | 6.2808 | 5.6769 | 5.4187 | 6.0621 | 5.2963 |
| GRB2 | 9.6572 | 8.8249 | 9.72 | 6.2743 | 6.3763 | 6.0937 | 6.9232 | 6.7712 |
| BRMS1 | 9.7466 | 9.7137 | 9.3484 | 7.2427 | 7.9508 | 6.9002 | 7.153 | 7.3846 |
| ARHGEF10 | 6.9846 | 7.408 | 6.1911 | 4.0145 | 3.6512 | 3.8089 | 3.7845 | 2.788 |
| WAC | 8.0588 | 7.144 | 7.5424 | 5.3401 | 5.0658 | 4.782 | 5.018 | 5.4298 |
| UBE2Z | 9.6449 | 9.2231 | 8.5817 | 6.5641 | 6.3528 | 6.3189 | 7.1393 | 6.6779 |
| TRMT112 | 13.7045 | 14.5817 | 13.5482 | 10.5427 | 10.7992 | 10.1815 | 10.1586 | 9.8995 |
| GOLPH3 | 9.1236 | 9.8097 | 8.8787 | 6.6094 | 6.9003 | 6.7482 | 7.028 | 6.9478 |
| PPP6R3 | 8.6388 | 8.8407 | 8.5095 | 6.5586 | 6.3713 | 6.1424 | 6.7132 | 6.2773 |
| EMC8 | 10.6656 | 10.3034 | 10.4708 | 8.7361 | 8.5856 | 8.5886 | 8.5008 | 8.7696 |
| RSAD1 | 7.0147 | 6.9352 | 6.446 | 4.4163 | 4.5152 | 4.3702 | 4.6876 | 4.89 |
| HAUS4 | 8.5911 | 8.6504 | 9.1966 | 5.9664 | 6.8442 | 5.8386 | 5.6776 | 6.1862 |
| TRIAP1 | 11.5136 | 11.2914 | 11.1692 | 8.4875 | 9.14 | 8.2346 | 8.6579 | 8.5847 |
| SMG8 | 9.0462 | 8.7764 | 8.9275 | 6.6147 | 6.099 | 6.8808 | 6.7126 | 7.108 |
| FAM13B | 7.9264 | 8.0175 | 7.8665 | 5.6468 | 5.895 | 6.1149 | 5.2588 | 5.6072 |
| PTRH2 | 12.0075 | 11.8882 | 11.9707 | 9.4661 | 9.3537 | 9.2109 | 9.1031 | 9.5597 |
| MTPAP | 8.7357 | 8.6081 | 9.1536 | 6.4507 | 5.7521 | 6.8745 | 5.8515 | 6.1696 |
| RRP15 | 10.531 | 11.0424 | 11.0715 | 8.4798 | 8.1431 | 8.6544 | 7.4609 | 7.7912 |
| TMEM185B | 6.6529 | 6.5491 | 6.531 | 4.7372 | 4.2591 | 4.602 | 4.3031 | 4.6284 |
| C7orf26 | 7.385 | 7.2374 | 7.3648 | 5.0961 | 5.1546 | 5.1655 | 5.6445 | 5.2983 |
| RBM15 | 7.5034 | 8.2485 | 7.2997 | 5.2196 | 5.4334 | 5.4418 | 4.5729 | 5.0905 |
| FA2H | 7.5278 | 7.8151 | 6.9056 | 3.9924 | 4.4871 | 3.7249 | 4.623 | 4.1229 |
| CHST11 | 6.04 | 5.6453 | 6.0121 | 3.5276 | 3.6529 | 3.7389 | 3.9195 | 3.0312 |
| RBM23 | 9.5494 | 9.099 | 9.1187 | 6.1562 | 7.1859 | 6.2482 | 6.4152 | 6.8197 |
| CNNM3 | 7.7045 | 7.3593 | 7.351 | 5.6699 | 5.3693 | 5.2071 | 5.8877 | 5.3843 |
| SOAT1 | 7.0557 | 7.4865 | 7.1193 | 4.7567 | 5.1418 | 4.661 | 4.8745 | 4.9938 |
| IMP3 | 9.722 | 9.6255 | 9.234 | 5.6594 | 5.7807 | 5.6287 | 5.9476 | 6.5707 |
| RNFT2 | 7.3289 | 7.8859 | 7.0099 | 4.4513 | 5.2512 | 4.1818 | 4.8003 | 4.5792 |
| CENPN | 7.8912 | 8.2269 | 7.9401 | 5.4073 | 5.3871 | 5.5317 | 5.7825 | 5.8112 |
| SLC25A44 | 8.3633 | 8.5898 | 8.1901 | 5.7512 | 5.9699 | 5.5315 | 6.1772 | 5.6301 |
KLF4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: KLF4
[1] Kruppel-like factor 4 (KLF4): What we currently know.
[2] KLF4 in Macrophages Attenuates TNFalpha-Mediated Kidney Injury and Fibrosis.
[3] The deubiquitinase USP10 regulates KLF4 stability and suppresses lung tumorigenesis.
[4] CD34(+)KLF4(+) Stromal Stem Cells Contribute to Endometrial Regeneration and Repair.
[8] ATXN3 promotes breast cancer metastasis by deubiquitinating KLF4.
[9] KLF4 as a rheostat of osteolysis and osteogenesis in prostate tumors in the bone.