CHD1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN1973
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: alvocidib
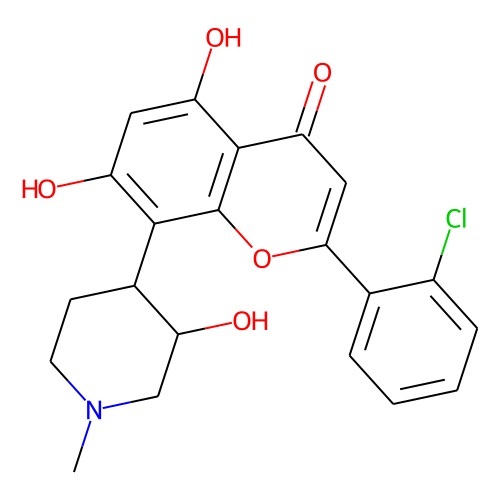
| ID | BRD-K87909389 |
| name | alvocidib |
| type | Small molecule |
| molecular formula | C21H20ClNO5 |
| molecular weight | 401.8 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | CN1CC[C@@H]([C@H](O)C1)c1c(O)cc(O)c2c1oc(cc2=O)-c1ccccc1Cl |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: CHD1
Transcription regulatory factors corresponding to this network
ARTICLE: alvocidib
[1] Overview of CDK9 as a target in cancer research.
[2] Transcription Elongation Can Affect Genome 3D Structure.
[4] Clinical activity of alvocidib (flavopiridol) in acute myeloid leukemia.
[5] YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers.
[6] Real-time dynamics of RNA polymerase II clustering in live human cells.
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CDK1 |
|
|
CDK2 
|
hsa04114 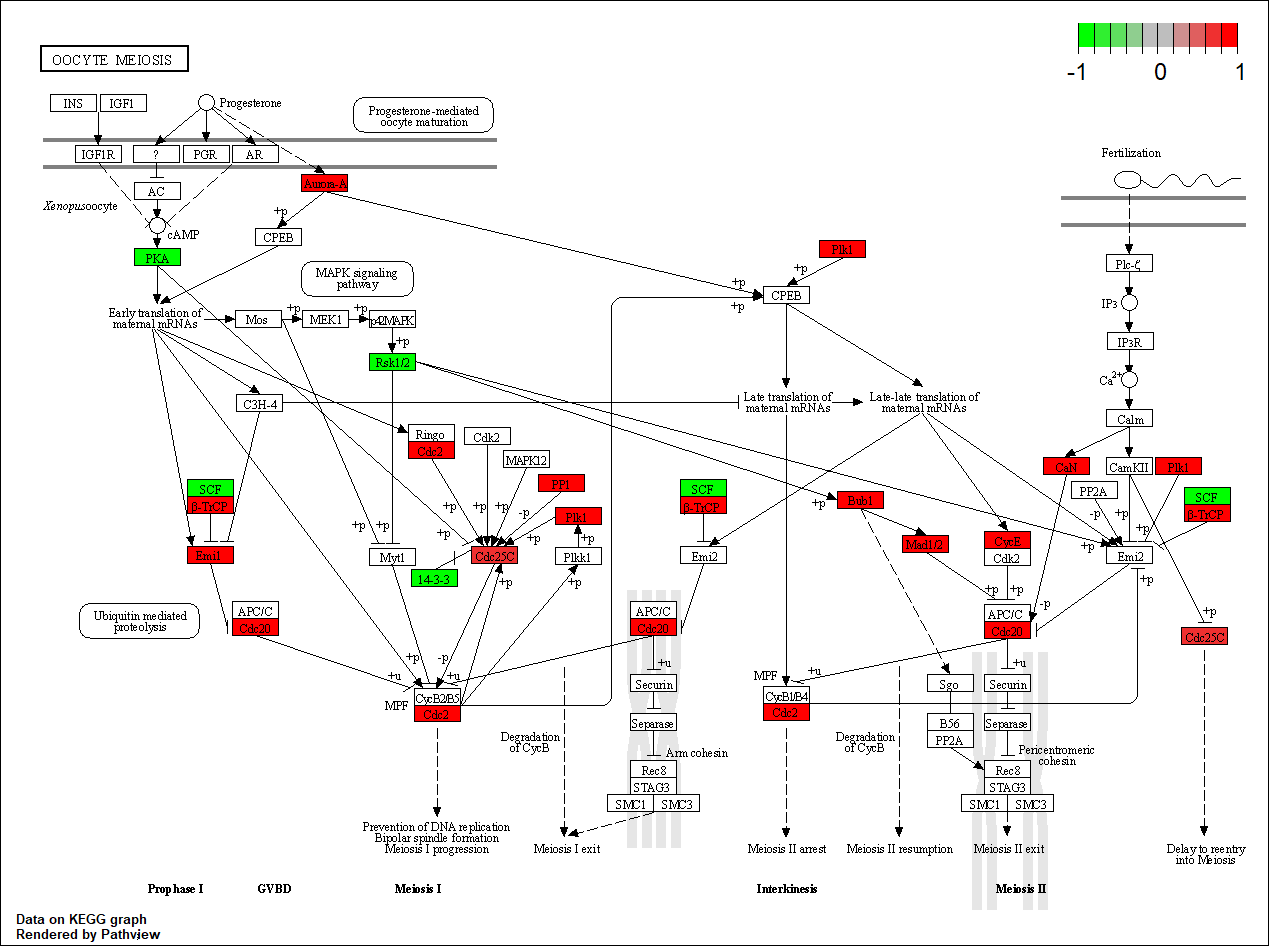
|
|
CDK4 |
hsa04115 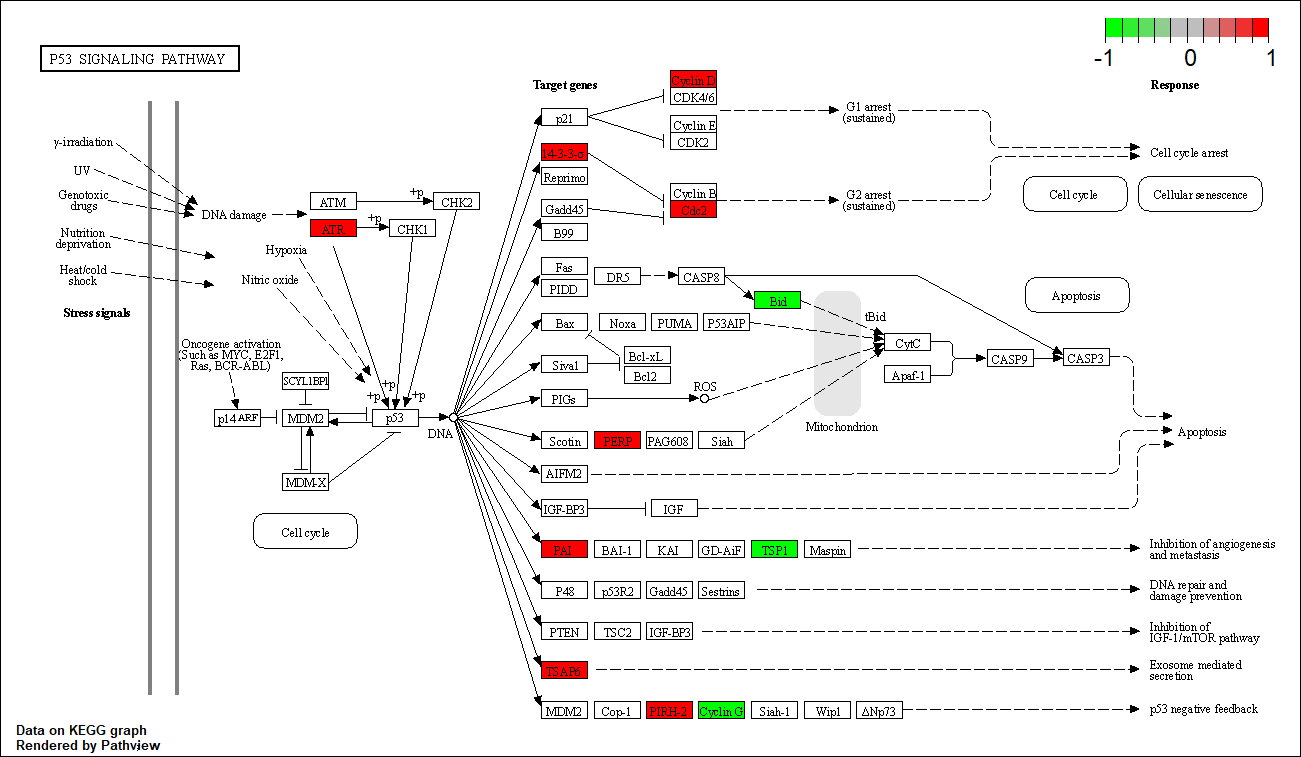
|
|
CDK5 
|
|
|
CDK6 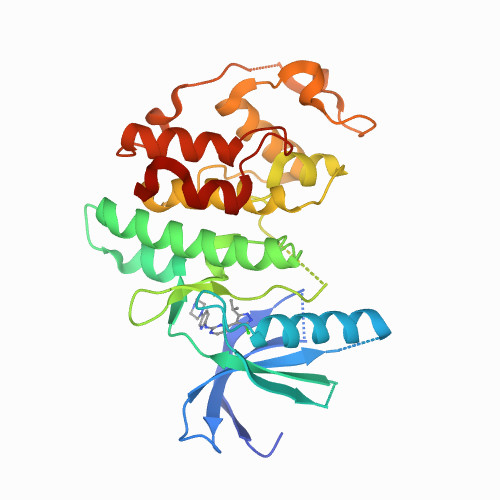
|
|
|
CDK7 
|
|
|
CDK8 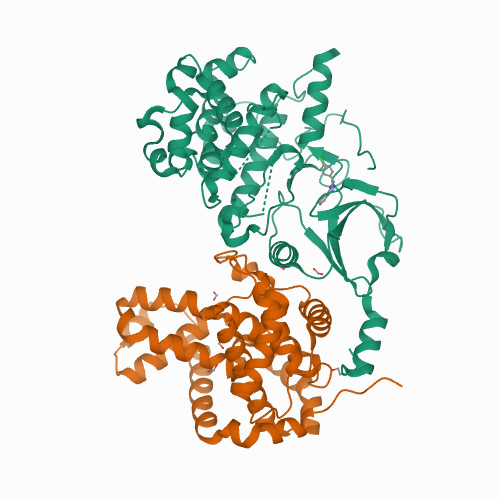
|
|
|
CDK9 
|
hsa05202 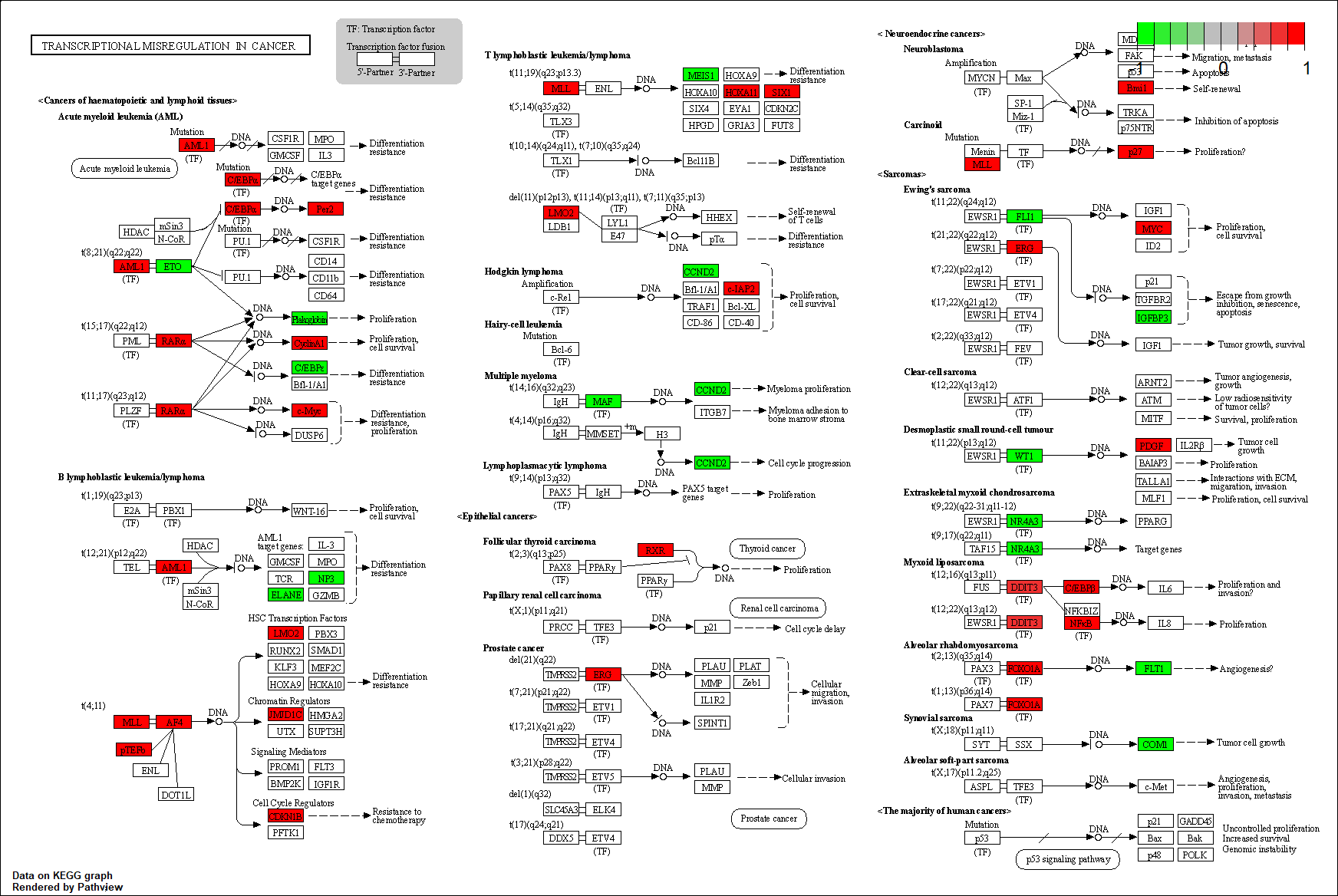
|
|
EGFR 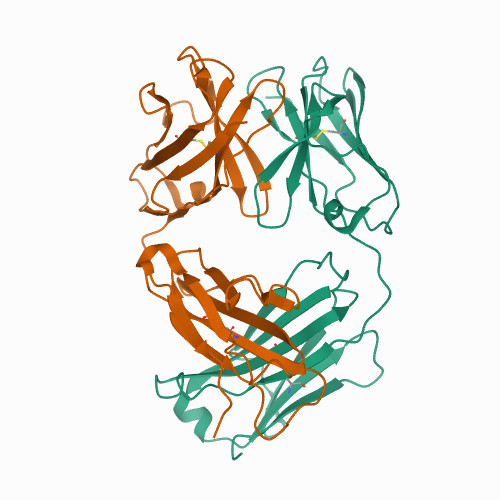
|
hsa04510 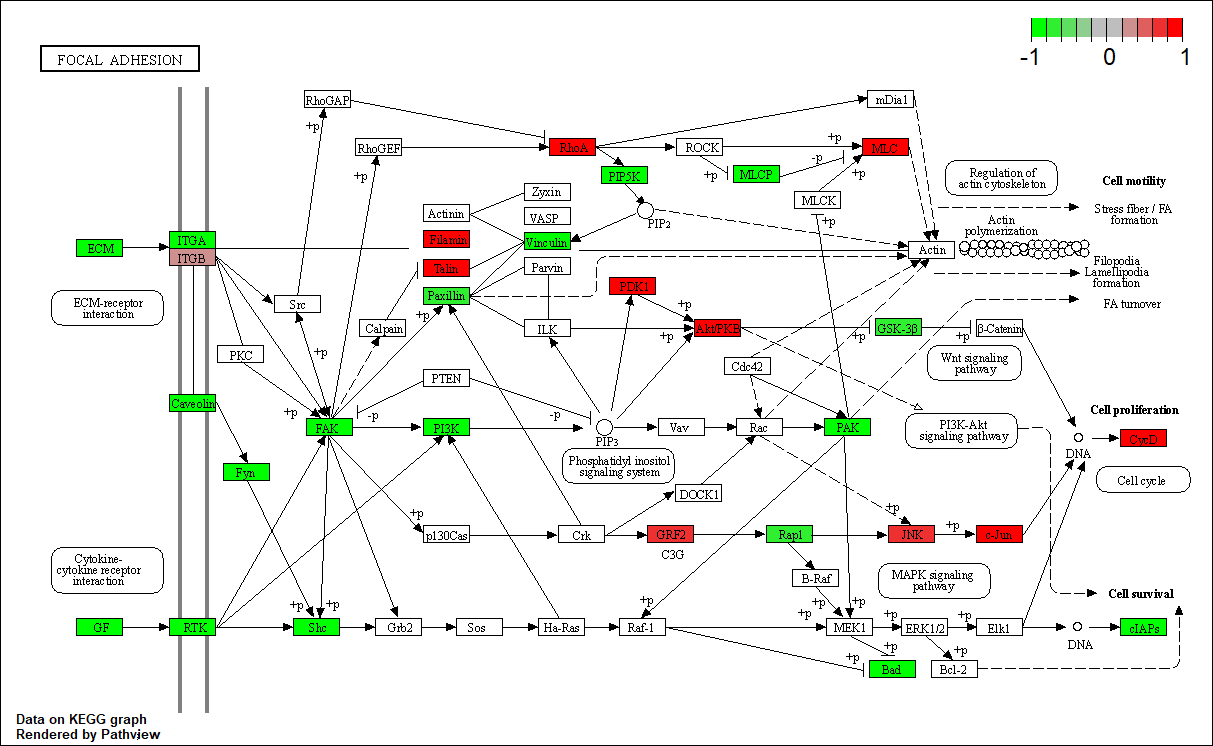
|
|
PYGM 
|
hsa04217 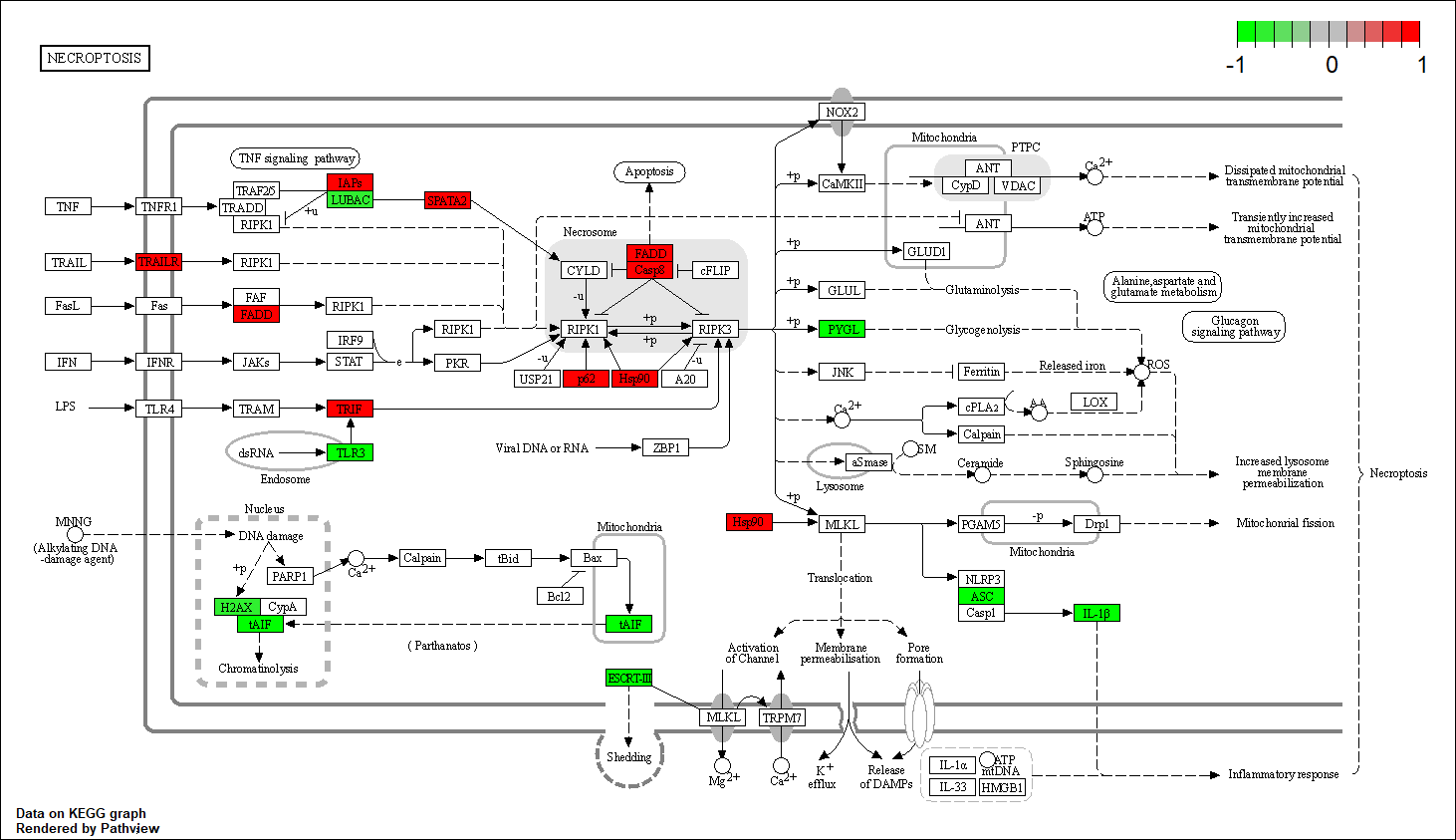
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|
| ZNF14 | 6.8726 | 7.7334 | 6.865 | 5.6997 | 5.8505 | 5.6971 | 5.3517 |
| CDC20 | 14.3523 | 13.9666 | 14.1011 | 12.2134 | 12.5494 | 12.4574 | 11.7441 |
| PLK1 | 11.2418 | 10.9259 | 11.4755 | 8.0663 | 4.6219 | 7.5331 | 6.7345 |
| TSKU | 7.8245 | 7.3839 | 6.4863 | 4.3934 | 3.6203 | 2.6822 | 3.217 |
| USP22 | 12.9228 | 12.6249 | 12.2309 | 10.4435 | 10.6961 | 10.4597 | 9.1273 |
| CHERP | 8.8945 | 8.9997 | 9.0079 | 6.1897 | 7.5718 | 7.0555 | 6.8696 |
| APPBP2 | 10.8577 | 10.7483 | 11.2114 | 9.319 | 9.9406 | 9.4341 | 9.0424 |
| TXLNA | 11.3263 | 11.2159 | 10.7546 | 8.3837 | 8.9102 | 8.4271 | 7.2267 |
| DNAJB1 | 11.1546 | 11.3751 | 10.4613 | 7.2535 | 7.2062 | 8.3802 | 6.5165 |
| USP1 | 7.4361 | 7.2653 | 7.1402 | 4.8907 | 5.5624 | 5.1673 | 5.2503 |
| KIF2C | 9.0936 | 9.3537 | 9.3101 | 7.6379 | 8.392 | 7.783 | 7.5769 |
| PLA2G15 | 7.2351 | 7.2879 | 7.3271 | 5.6606 | 6.117 | 5.4689 | 4.8802 |
| PIP4K2B | 5.4166 | 5.1816 | 5.2389 | 3.5349 | 3.9498 | 3.7874 | 4.1972 |
| CYTH1 | 5.9477 | 5.9246 | 5.4106 | 4.7891 | 4.506 | 4.2693 | 4.4595 |
| KEAP1 | 10.5292 | 10.6866 | 10.1916 | 8.2011 | 8.8054 | 8.84 | 8.8715 |
| BAMBI | 11.7051 | 11.6528 | 11.7377 | 8.5129 | 7.5297 | 7.3996 | 6.0754 |
| RNMT | 7.7404 | 7.8105 | 7.7334 | 6.095 | 6.1863 | 5.1673 | 5.1005 |
| NET1 | 9.1718 | 10.0892 | 9.9949 | 7.6448 | 7.5718 | 7.3116 | 7.766 |
| SCYL3 | 6.1168 | 5.3067 | 4.5305 | 3.4171 | 3.2475 | 2.4668 | 3.4503 |
| JMJD6 | 11.5627 | 11.3968 | 10.7992 | 8.2104 | 9.0252 | 8.4852 | 7.3765 |
| CDCA4 | 7.4219 | 8.8784 | 7.9946 | 4.5422 | 5.7982 | 5.0449 | 5.1196 |
| CANT1 | 12.4553 | 12.2577 | 12.3474 | 10.7089 | 11.1473 | 11.2296 | 10.9693 |
| EIF5 | 10.9083 | 11.1822 | 11.6026 | 8.2104 | 8.8258 | 8.5947 | 9.4304 |
| GATA3 | 10.8233 | 10.7866 | 10.5649 | 6.6082 | 6.1613 | 6.1943 | 5.3653 |
| TIMM9 | 13.6579 | 13.3626 | 13.8636 | 10.755 | 10.6147 | 10.6107 | 10.7149 |
| PARP2 | 9.1153 | 8.9714 | 8.9543 | 6.4365 | 4.3204 | 5.7471 | 5.3498 |
| RPP38 | 7.9203 | 7.7192 | 8.9543 | 4.8265 | 5.5053 | 3.8366 | 4.7563 |
| BAG3 | 12.525 | 12.6861 | 13.09 | 9.779 | 9.9356 | 8.4852 | 9.4891 |
| DLGAP5 | 10.0186 | 10.0284 | 9.7563 | 8.3351 | 8.4158 | 7.9005 | 7.8653 |
| ZC3H4 | 8.0467 | 7.7184 | 7.9032 | 6.1909 | 5.7233 | 6.3455 | 5.6345 |
| ZNF146 | 9.5066 | 9.2572 | 9.2738 | 8.2233 | 8.465 | 7.6891 | 7.7466 |
| IST1 | 10.501 | 10.6238 | 10.5775 | 9.0563 | 7.1644 | 8.5005 | 8.7182 |
| DDX39A | 13.7218 | 13.8046 | 13.5877 | 10.6251 | 10.5022 | 12.3205 | 11.1555 |
| PSMC6 | 12.5494 | 13.0815 | 12.4919 | 11.5082 | 10.4945 | 10.8392 | 10.8814 |
| VEZF1 | 9.5148 | 8.8014 | 8.048 | 6.772 | 7.1596 | 6.6225 | 6.4502 |
| TRAF4 | 8.3711 | 8.3687 | 8.6413 | 6.5391 | 6.7009 | 7.5313 | 6.5252 |
| CREBZF | 7.5477 | 7.6317 | 7.4507 | 6.2605 | 6.6002 | 6.4116 | 5.7655 |
| BAG5 | 8.3345 | 9.1791 | 8.5537 | 6.2248 | 5.4128 | 6.7969 | 6.1098 |
| CHSY1 | 6.8076 | 6.7304 | 6.5722 | 4.7765 | 3.3043 | 4.4284 | 4.1371 |
| TFAM | 10.2298 | 10.703 | 10.2922 | 9.1645 | 8.7951 | 8.0338 | 8.6586 |
| PCF11 | 6.0991 | 6.6875 | 6.7425 | 4.2755 | 4.839 | 4.8001 | 4.567 |
| MED1 | 7.9799 | 8.0079 | 7.875 | 5.8087 | 6.1152 | 6.6091 | 6.2539 |
| FSTL3 | 7.5917 | 6.649 | 6.9991 | 5.4453 | 5.4838 | 5.5662 | 5.5303 |
| COIL | 9.6544 | 9.5349 | 9.3802 | 7.8939 | 7.4502 | 7.3153 | 7.2789 |
| TDG | 11.1961 | 12.2183 | 11.3124 | 10.0741 | 8.6129 | 9.6677 | 9.6134 |
| JUND | 11.844 | 12.3936 | 12.8628 | 10.2784 | 9.8577 | 10.7353 | 10.5614 |
| TOE1 | 7.5614 | 7.4507 | 7.4014 | 6.3129 | 6.2223 | 6.4786 | 6.5943 |
| RFC3 | 11.3938 | 12.3776 | 12.0144 | 10.1382 | 9.7618 | 10.0949 | 10.1317 |
| USPL1 | 6.8792 | 7.5898 | 7.0427 | 5.3761 | 5.3025 | 5.7784 | 5.0506 |
| ZNF202 | 6.3969 | 6.9313 | 6.4114 | 5.071 | 5.0095 | 5.4169 | 5.5936 |
| CEP350 | 6.7353 | 7.0337 | 6.6128 | 4.4896 | 4.239 | 5.1116 | 5.1304 |
| FEN1 | 11.9447 | 12.7412 | 11.7315 | 10.5212 | 10.111 | 10.8701 | 10.575 |
| URB2 | 7.1852 | 7.3498 | 6.8636 | 5.4091 | 5.0426 | 5.8594 | 5.5311 |
| GEMIN4 | 8.7593 | 9.3188 | 9.2196 | 7.2441 | 7.2645 | 7.0587 | 7.0897 |
| ALKBH1 | 7.1605 | 7.2771 | 7.694 | 5.1695 | 4.4895 | 5.6795 | 5.2692 |
| BLM | 6.1173 | 6.2683 | 5.3061 | 4.0787 | 4.4132 | 3.583 | 4.2819 |
| ZNF443 | 6.7547 | 6.3318 | 5.8371 | 4.571 | 4.7599 | 4.0845 | 4.088 |
| HSPH1 | 12.6312 | 12.0542 | 13.6198 | 10.7089 | 10.0887 | 10.8471 | 10.2922 |
| ZNF200 | 6.3639 | 6.4752 | 6.233 | 4.7965 | 4.6591 | 5.0702 | 5.0574 |
| ARHGAP12 | 5.8114 | 6.7989 | 6.516 | 4.6123 | 4.5013 | 4.1343 | 4.5759 |
| SLC19A2 | 8.4988 | 9.1325 | 8.2603 | 5.6167 | 5.897 | 5.9834 | 4.1395 |
| TAF5 | 7.425 | 8.1789 | 7.1442 | 5.9196 | 5.9959 | 5.4409 | 5.8594 |
| TUBD1 | 10.7483 | 10.6722 | 10.2397 | 8.8998 | 8.6075 | 8.5757 | 7.9318 |
| FBL | 12.1775 | 11.8806 | 12.3658 | 11.125 | 10.602 | 10.9767 | 10.9386 |
| KPNA2 | 14.3049 | 15 | 13.7901 | 12.511 | 12.0978 | 12.5949 | 11.4408 |
| PNN | 11.0636 | 10.8619 | 11.5772 | 9.8907 | 9.6134 | 9.4147 | 8.6507 |
| SLC7A1 | 12.9813 | 12.8189 | 12.268 | 10.4579 | 10.7205 | 9.337 | 8.2784 |
| FEM1B | 8.7598 | 9.4565 | 9.3967 | 7.8059 | 7.6166 | 7.7766 | 7.8418 |
| ELF1 | 8.3108 | 7.9762 | 8.522 | 6.2786 | 4.9944 | 6.32 | 5.2161 |
| MFSD5 | 9.158 | 9.6595 | 9.207 | 8.0742 | 7.6825 | 8.0842 | 7.8932 |
| ZFC3H1 | 7.6625 | 7.5633 | 7.8209 | 6.3456 | 6.7455 | 6.6462 | 6.1014 |
| ZBTB1 | 8.2011 | 7.4487 | 7.5328 | 5.1735 | 4.7625 | 5.0443 | 5.4912 |
| EDRF1 | 6.9282 | 7.5461 | 6.6823 | 5.0176 | 5.081 | 4.9884 | 5.1474 |
| METTL18 | 7.0359 | 7.2168 | 7.2291 | 5.1025 | 4.9008 | 4.6789 | 5.8709 |
| NGDN | 10.8619 | 10.8006 | 11.0802 | 8.9085 | 8.4051 | 8.6438 | 8.8168 |
| CA12 | 9.9908 | 11.6407 | 10.6069 | 8.9163 | 7.8787 | 8.7326 | 7.8896 |
| HNRNPA1 | 11.9901 | 11.8374 | 11.6439 | 10.597 | 9.6808 | 9.5827 | 9.6868 |
| PIAS1 | 6.6424 | 7.4552 | 7.0567 | 5.7612 | 5.5844 | 5.346 | 5.8798 |
| BAZ1A | 9.5869 | 9.5949 | 9.3946 | 7.5641 | 7.0252 | 7.9454 | 7.3042 |
| GTPBP4 | 11.1147 | 11.7455 | 11.4305 | 9.6322 | 9.6858 | 8.8849 | 9.4873 |
| ZBED5 | 7.7593 | 7.5137 | 7.7085 | 6.162 | 6.1627 | 6.1677 | 5.9994 |
| SAV1 | 7.709 | 7.3572 | 7.7397 | 5.9863 | 5.8001 | 5.9968 | 5.2372 |
| S100PBP | 6.7007 | 7.5589 | 7.0187 | 5.1617 | 5.0599 | 5.3086 | 4.9687 |
| PSPC1 | 8.0163 | 7.688 | 8.1404 | 6.233 | 5.4711 | 6.6879 | 6.0285 |
| ZCCHC8 | 9.4965 | 10.2022 | 9.107 | 7.9838 | 7.9679 | 8.2573 | 7.5803 |
| SMG8 | 8.7837 | 8.7454 | 8.5911 | 7.2051 | 7.6304 | 6.9934 | 6.7852 |
| IER5 | 10.448 | 11.9212 | 10.9298 | 8.4277 | 9.1835 | 8.4573 | 9.1258 |
| PTRH2 | 11.4669 | 12.0073 | 11.6471 | 9.3639 | 10.145 | 9.1028 | 10.1916 |
| TAF1D | 7.2321 | 7.4488 | 7.507 | 5.6541 | 5.39 | 5.2151 | 5.7567 |
| RNF111 | 5.5539 | 5.507 | 5.6055 | 2.9219 | 4.0441 | 3.9323 | 4.0725 |
| ZNF574 | 7.5033 | 7.5341 | 8.0961 | 6.1179 | 6.0013 | 6.7792 | 5.9707 |
| SIRT1 | 6.2323 | 7.3981 | 6.7923 | 4.903 | 5.5469 | 4.9686 | 4.7527 |
| MTPAP | 8.3385 | 8.9867 | 8.4821 | 6.8706 | 6.9816 | 6.7329 | 7.155 |
| ZNHIT2 | 7.4371 | 7.4539 | 7.0718 | 6.1776 | 6.216 | 5.8138 | 6.1077 |
| ZCCHC2 | 6.8516 | 6.377 | 7.6552 | 5.0804 | 5.4033 | 5.4434 | 4.9734 |
| SPHK1 | 7.3686 | 6.8656 | 7.8602 | 5.156 | 4.9955 | 4.7087 | 5.6605 |
| RBM15 | 7.7364 | 7.7897 | 8.1634 | 5.3641 | 6.6236 | 5.3529 | 4.8936 |
| NAA16 | 8.8006 | 9.313 | 9.6467 | 8.0297 | 7.7867 | 8.0423 | 7.7253 |
| PALB2 | 6.6951 | 7.2713 | 7.2912 | 5.1381 | 3.9314 | 4.645 | 5.3633 |
| ZNF267 | 5.8649 | 6.2512 | 5.9335 | 4.6418 | 4.1375 | 3.5647 | 4.2534 |
| TDP1 | 7.742 | 7.6681 | 7.4162 | 5.946 | 4.2778 | 5.8034 | 4.7423 |
| MCM10 | 8.8483 | 8.8818 | 9.6321 | 7.0916 | 7.1905 | 6.5958 | 6.994 |
| MRTO4 | 11.5117 | 11.6829 | 11.5105 | 10.5368 | 10.354 | 9.8856 | 10.5817 |
| C1orf112 | 7.5665 | 7.9939 | 7.6563 | 6.5017 | 5.9914 | 6.3846 | 6.8074 |
| CDCA8 | 11.304 | 11.2222 | 10.9646 | 8.9426 | 9.4865 | 9.7875 | 9.9416 |
| C11orf68 | 6.9684 | 6.9409 | 6.7409 | 5.8041 | 5.7003 | 5.6952 | 5.0909 |
| USP3 | 10.4794 | 11.7159 | 11.9599 | 9.5645 | 9.2572 | 9.3851 | 9.236 |
| IMP3 | 9.4169 | 9.3821 | 8.9467 | 7.5788 | 7.6392 | 7.1753 | 6.0247 |
| BRIP1 | 8.8512 | 8.0197 | 8.2171 | 7.1505 | 7.0698 | 6.8686 | 6.5237 |
| NOL11 | 13.3998 | 15 | 13.6141 | 11.6909 | 11.553 | 11.5867 | 11.9098 |
| DCLRE1C | 6.1843 | 6.1614 | 5.1963 | 3.5496 | 4.1035 | 2.6543 | 3.7365 |
| SLC25A44 | 7.981 | 7.9381 | 7.918 | 6.9012 | 6.172 | 6.3248 | 5.9912 |
| CEP76 | 6.7952 | 6.7849 | 6.0253 | 4.7804 | 5.2824 | 4.8677 | 4.5574 |
CHD1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: CHD1
[1] SPOP-Mutated/CHD1-Deleted Lethal Prostate Cancer and Abiraterone Sensitivity.
[3] Nucleosome-Chd1 structure and implications for chromatin remodelling.
[4] Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer.
[9] The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans.
[10] CHD1 and SPOP synergistically protect prostate epithelial cells from DNA damage.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below