PTPN11 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN2027
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: dovitinib
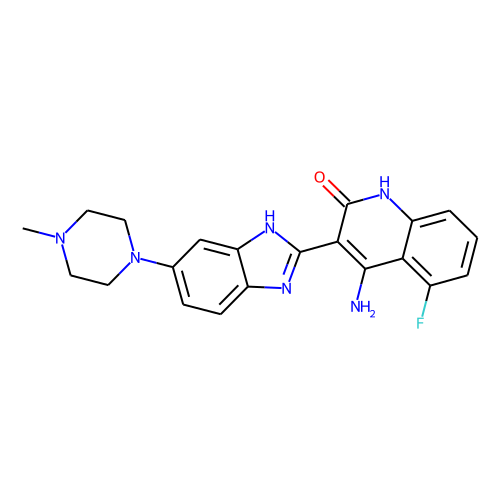
| ID | BRD-K85402309 |
| name | dovitinib |
| type | Small molecule |
| molecular formula | C21H21FN6O |
| molecular weight | 392.4 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | CN1CCN(CC1)c1ccc2nc([nH]c2c1)-c1c(N)c2c(F)cccc2[nH]c1=O |
CONDITION TYPE: trt_cp
Compound
CELL LINE: A549
Cell lines used for experiments
TRANSCRIPTION FACTOR: PTPN11
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CSF1R 
|
hsa05202 
|
|
EGFR 
|
hsa04520 
|
|
FGFR1 |
hsa04015 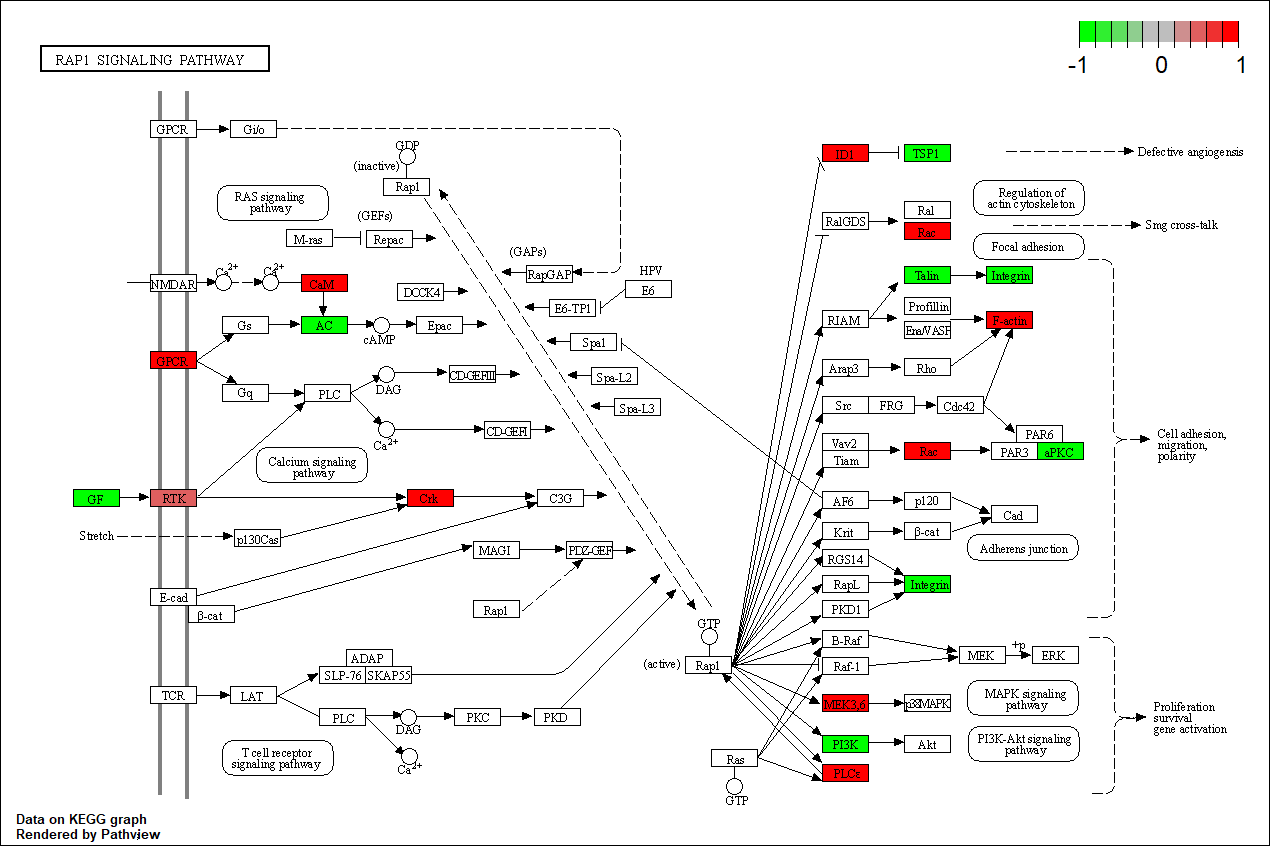
|
|
FGFR2 
|
hsa04144 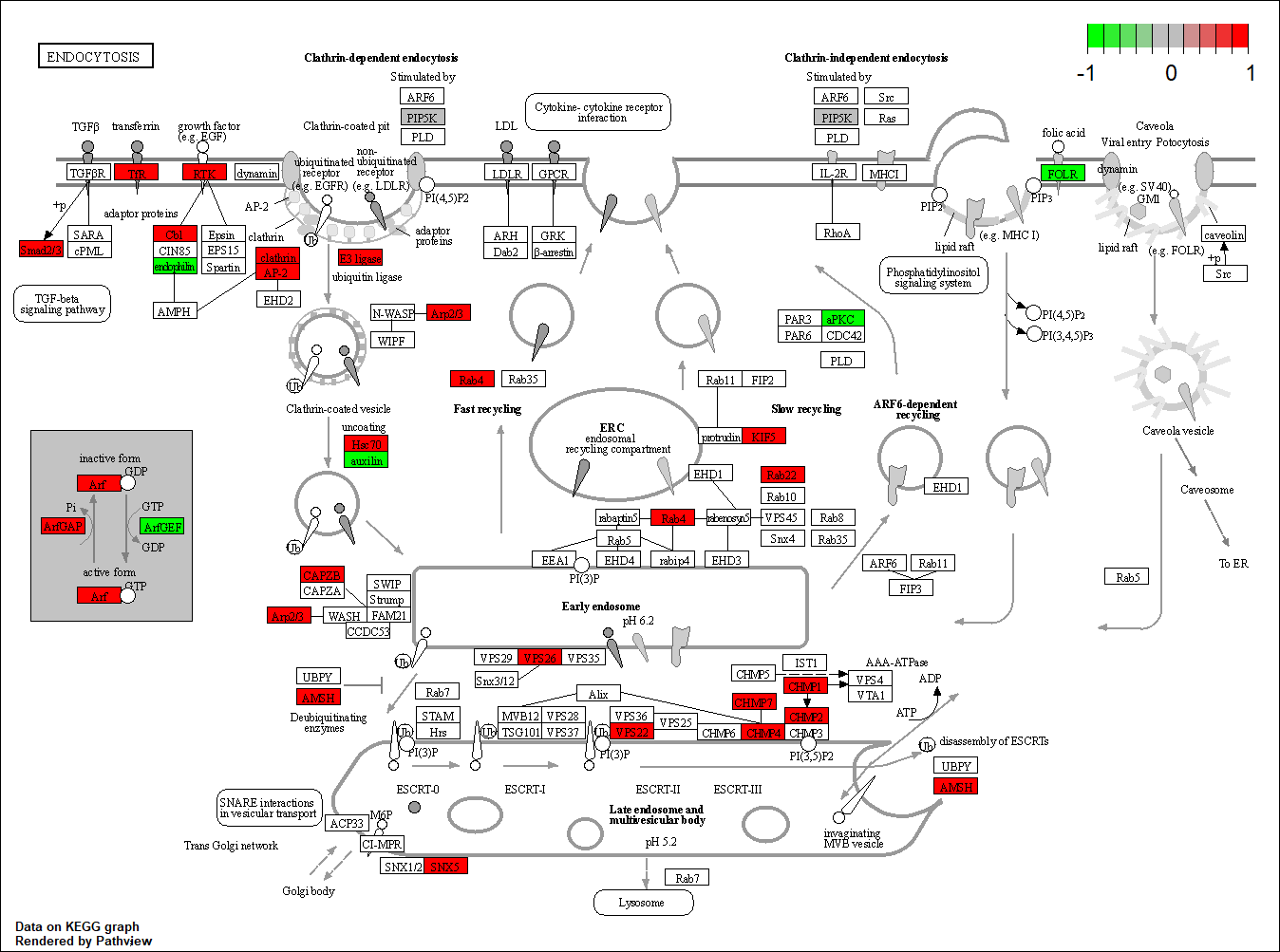
|
|
FGFR3 |
hsa05230 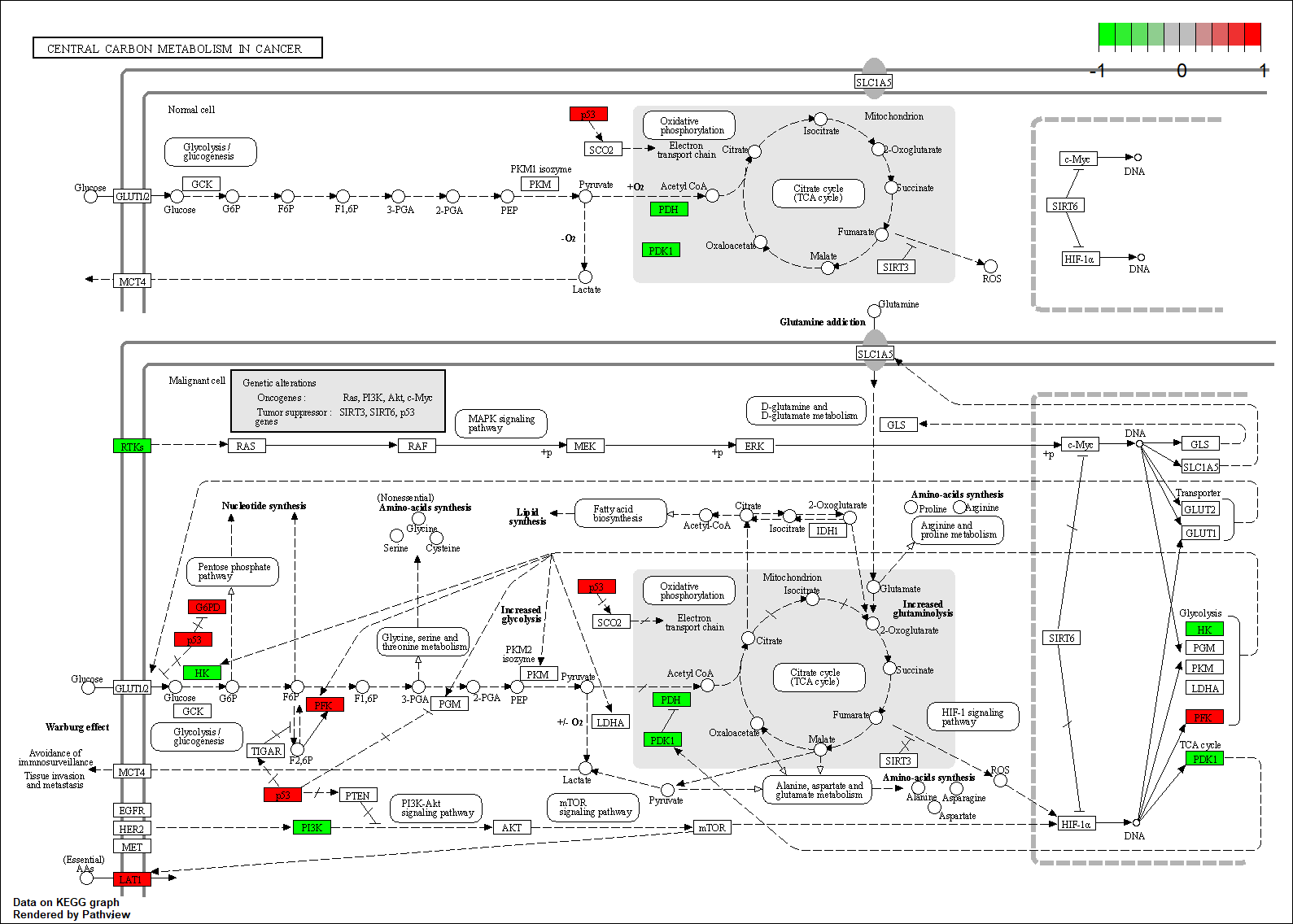
|
|
FLT1 
|
hsa04010 
|
|
FLT3 
|
hsa05230 
|
|
FLT4 |
hsa04151 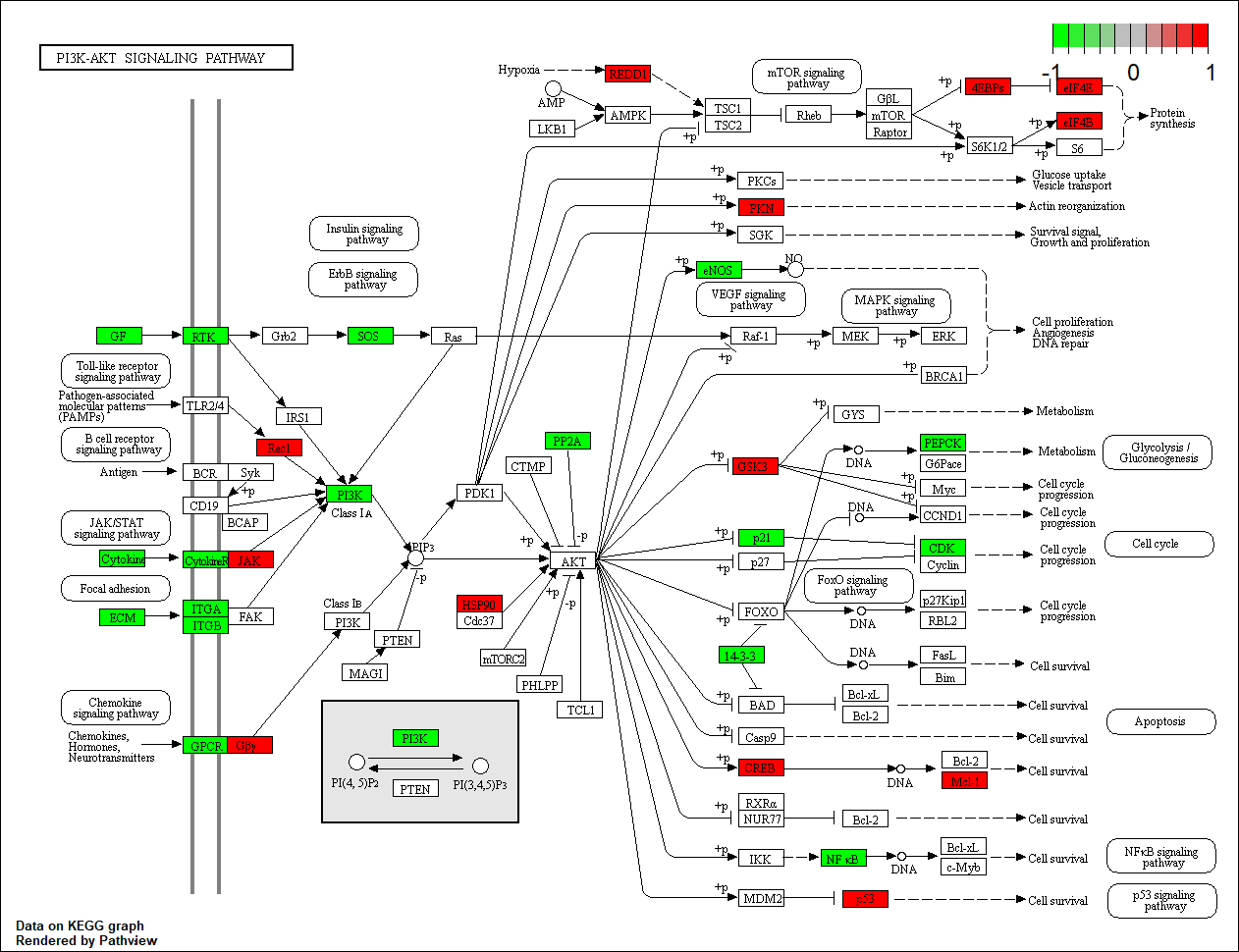
|
|
INSR 
|
hsa04010 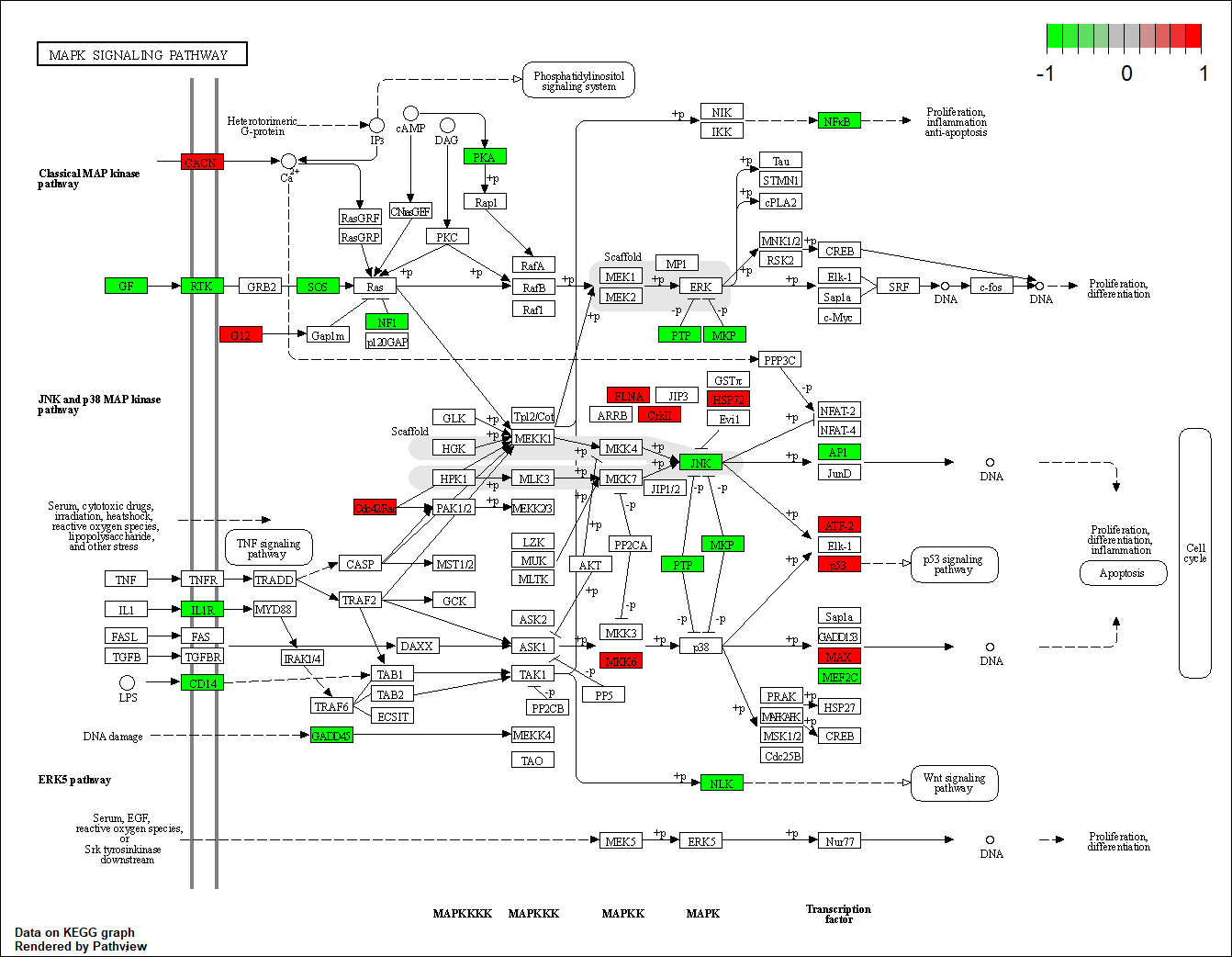
|
|
KDR 
|
|
|
KIT 
|
hsa04640 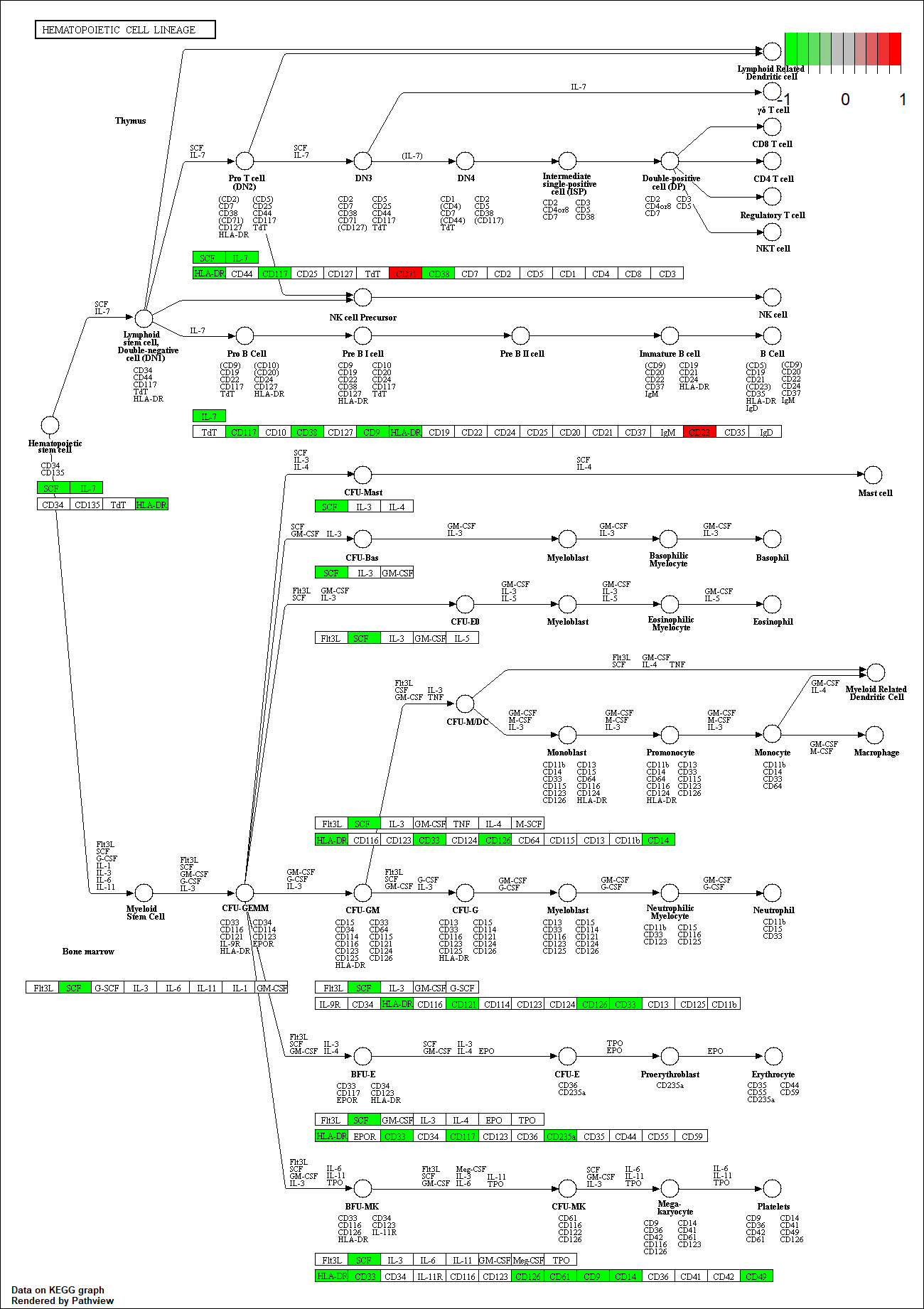
|
|
PDGFRA 
|
hsa04144 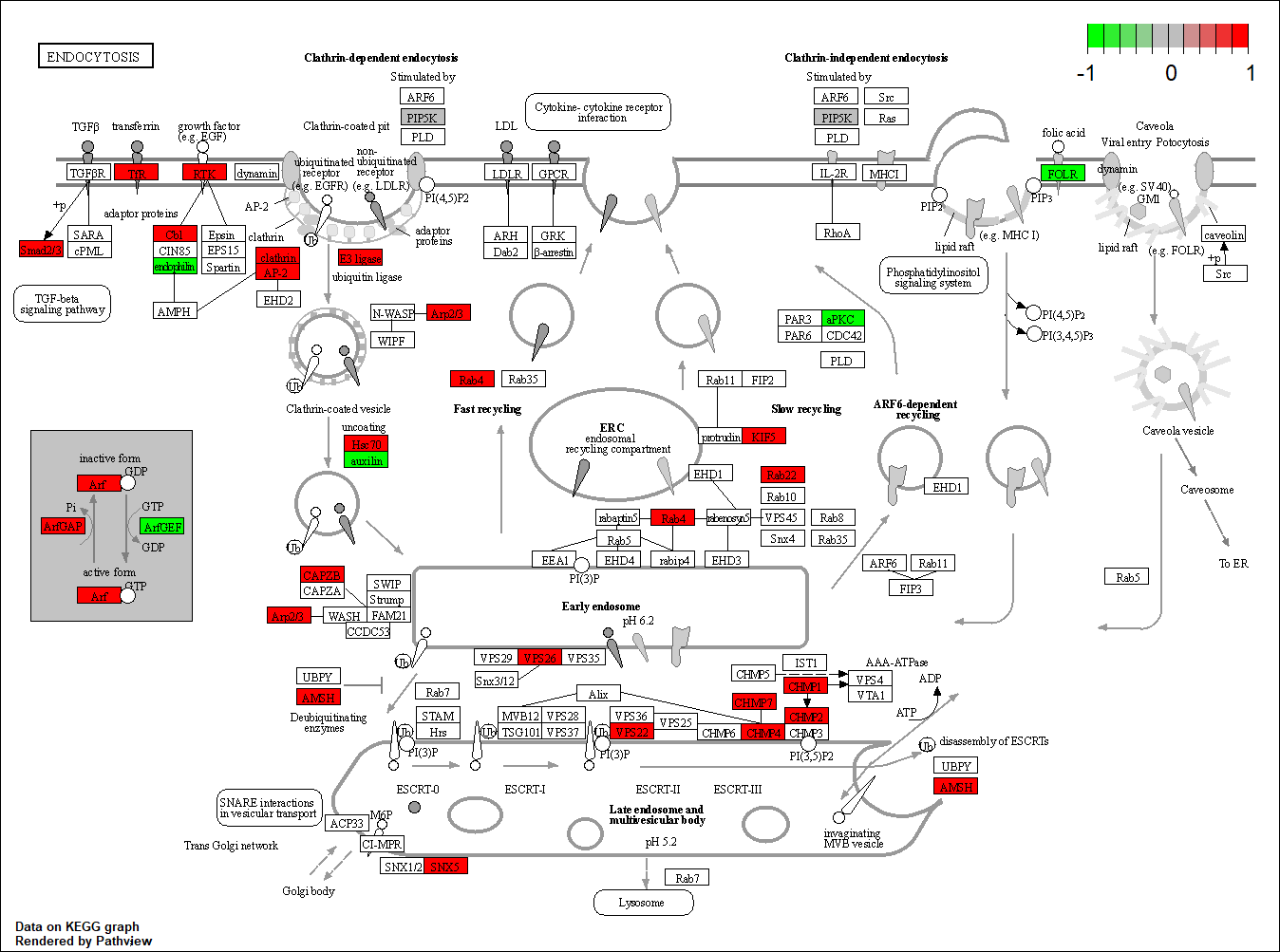
|
|
PDGFRB 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| C1orf54 | 9.2531 | 8.7229 | 8.6701 | 6.8251 | 4.9592 | 5.2279 |
| ACTB | 13.6613 | 13.5866 | 13.4338 | 10.3932 | 11.9844 | 10.4804 |
| HSPD1 | 10.8076 | 11.7964 | 12.2848 | 5.4359 | 9.0695 | 7.5819 |
| MNAT1 | 11.0803 | 10.6959 | 11.5205 | 8.8634 | 9.5037 | 9.1084 |
| PAK4 | 9.3062 | 9.8656 | 9.4216 | 6.6362 | 5.6008 | 4.2653 |
| PAICS | 5.6495 | 6.3727 | 6.8585 | 4.3974 | 3.537 | 3.2584 |
| SLC35B1 | 11.1667 | 12.8414 | 11.9317 | 10.0616 | 7.7846 | 7.5206 |
| CDK1 | 12.3113 | 12.1562 | 11.6924 | 10.1557 | 9.271 | 9.6751 |
| ZMYM2 | 9.1049 | 9.9764 | 9.4484 | 6.2959 | 4.8885 | 4.6096 |
| PSIP1 | 8.0557 | 8.3364 | 9.0398 | 3.6851 | 4.4281 | 4.2003 |
| HSD17B11 | 11.1732 | 11.0422 | 11.4488 | 8.3696 | 4.6797 | 6.6561 |
| PECR | 7.5774 | 7.9457 | 8.1047 | 6.1988 | 4.2092 | 4.3916 |
| TBC1D31 | 10.213 | 10.6646 | 10.9258 | 8.6084 | 6.7564 | 7.4995 |
| ARID4B | 7.2143 | 8.2165 | 7.3612 | 3.7652 | 4.8885 | 5.8488 |
| RAB31 | 10.2014 | 9.2849 | 9.5918 | 7.1838 | 5.1592 | 7.4047 |
| DLGAP5 | 9.0952 | 8.8276 | 10.2792 | 6.4495 | 5.5382 | 7.3817 |
| SNRPF | 13.7973 | 14.161 | 13.5952 | 10.3119 | 9.3526 | 9.5325 |
| CBX3 | 11.6275 | 10.1058 | 11.3703 | 6.9828 | 8.7624 | 7.4037 |
| C1D | 10.7001 | 9.7015 | 10.8156 | 8.3172 | 7.099 | 8.3611 |
| TMED2 | 11.1934 | 11.6166 | 12.0159 | 9.4362 | 9.5023 | 8.1316 |
| HNRNPK | 11.4673 | 11.5185 | 11.0487 | 9.3012 | 9.5521 | 8.5384 |
| PHB | 13.6043 | 14.4107 | 14.0136 | 11.8085 | 9.5932 | 9.4721 |
| BZW1 | 10.3874 | 10.8153 | 10.8538 | 7.7925 | 6.4102 | 6.4306 |
| FUS | 11.129 | 11.9726 | 10.5541 | 7.7998 | 7.0765 | 7.0076 |
| SNX17 | 11.1399 | 11.4404 | 10.3609 | 8.3719 | 8.4714 | 8.5032 |
| PSMA7 | 15 | 15 | 15 | 11.3115 | 12.6174 | 10.8261 |
| GSS | 11.705 | 12.8503 | 12.2153 | 8.0798 | 8.2003 | 9.461 |
| NQO1 | 14.0788 | 13.1856 | 15 | 9.9972 | 11.7584 | 11.1262 |
| RCN2 | 7.9903 | 7.7101 | 8.7341 | 4.7228 | 6.3342 | 4.2876 |
| PSMA3 | 13.3693 | 12.9864 | 14.0367 | 11.4419 | 10.2775 | 10.5506 |
| MARCKS | 11.6387 | 11.3463 | 13.0885 | 7.1417 | 6.0545 | 8.1184 |
| ABCE1 | 9.2257 | 8.6307 | 10.0567 | 7.5127 | 5.9714 | 5.4121 |
| PON2 | 11.4595 | 10.3584 | 10.8153 | 4.5694 | 2.578 | 6.1672 |
| N4BP2L2 | 7.6287 | 6.7361 | 7.6298 | 5.4459 | 5.2821 | 5.0027 |
| SREBF1 | 10.1902 | 10.4154 | 11.0305 | 5.7958 | 7.3446 | 8.082 |
| MTHFD1 | 12.333 | 12.534 | 12.1049 | 9.8855 | 6.4592 | 7.7484 |
| HSPBP1 | 11.9533 | 13.0156 | 11.5347 | 9.4067 | 8.6058 | 10.2171 |
| DNAJC7 | 13.2062 | 14.2241 | 15 | 10.618 | 8.4691 | 10.694 |
| SSBP1 | 11.9361 | 12.3836 | 12.761 | 9.2973 | 8.9624 | 10.5534 |
| TOB1 | 8.522 | 8.8319 | 8.532 | 4.8073 | 6.3987 | 5.1445 |
| UBE2S | 13.9528 | 15 | 13.4825 | 7.2792 | 6.99 | 10.2037 |
| AAGAB | 10.3626 | 10.9493 | 10.6406 | 7.0295 | 6.9228 | 7.4389 |
| NBN | 9.7397 | 9.3703 | 9.6745 | 6.4394 | 8.0979 | 6.8568 |
| RNASEH2A | 11.1159 | 11.7432 | 10.3392 | 9.197 | 8.2041 | 8.3947 |
| UROS | 9.2987 | 9.6705 | 9.4326 | 7.2378 | 7.9092 | 7.4627 |
| ARHGEF1 | 10.2045 | 10.3574 | 9.3227 | 6.8959 | 7.563 | 7.4902 |
| SPAG5 | 9.8449 | 9.0224 | 9.4653 | 7.0347 | 7.8154 | 6.2429 |
| NDUFB3 | 10.6423 | 10.9651 | 10.0777 | 8.1798 | 8.4788 | 6.9815 |
| PSMA4 | 12.3638 | 11.7274 | 11.7892 | 10.0321 | 7.9949 | 9.5921 |
| PTTG1 | 10.7531 | 12.4188 | 11.5809 | 8.8778 | 6.147 | 8.294 |
| DYNC1LI2 | 12.5253 | 11.6252 | 11.6973 | 9.1694 | 5.7309 | 8.4982 |
| EMP3 | 9.1514 | 10.6924 | 11.4688 | 6.6776 | 7.6047 | 5.8721 |
| TDG | 9.1987 | 9.2993 | 10.2972 | 6.3289 | 6.6291 | 7.6312 |
| CEACAM6 | 4.3626 | 4.2428 | 5.0693 | 0.9866 | 1.2836 | 0.3535 |
| CSTF3 | 8.6259 | 8.68 | 8.8893 | 5.9903 | 4.9169 | 4.4254 |
| PPIH | 9.4179 | 9.5149 | 10.0954 | 7.3815 | 7.1361 | 7.7131 |
| SLC12A2 | 10.4368 | 11.1273 | 9.0601 | 7.4753 | 7.8047 | 7.1892 |
| TOM1L1 | 9.9157 | 10.3864 | 9.5389 | 6.6718 | 4.3281 | 3.4745 |
| PDE4D | 8.6618 | 8.0491 | 9.1425 | 4.6734 | 4.267 | 6.5567 |
| NAP1L1 | 11.9966 | 11.2158 | 12.6823 | 9.118 | 7.2311 | 7.8679 |
| MLF1 | 7.7869 | 9.1642 | 9.3901 | 4.6792 | 6.638 | 5.1328 |
| POP5 | 11.2628 | 10.8809 | 11.5231 | 8.8246 | 8.4774 | 9.4581 |
| BCL3 | 9.6401 | 10.6476 | 9.551 | 6.5008 | 7.1967 | 6.2362 |
| PRIM1 | 9.0746 | 9.2877 | 8.8062 | 6.547 | 7.1566 | 7.0554 |
| HNRNPA2B1 | 12.9799 | 12.7039 | 13.3461 | 10.9947 | 11.2724 | 10.5191 |
| IGFBP1 | 7.8168 | 7.6374 | 7.343 | 5.0335 | 4.8609 | 4.3626 |
| SNAPC1 | 8.8474 | 8.292 | 8.4748 | 5.7941 | 5.6519 | 3.2492 |
| RBM42 | 12.0986 | 13.1835 | 11.1914 | 9.8797 | 9.0516 | 8.7675 |
| AKAP7 | 8.0915 | 7.4091 | 8.5109 | 3.1717 | 5.7211 | 2.1975 |
| SIX1 | 9.6527 | 8.6589 | 8.4675 | 6.0753 | 6.7877 | 5.6068 |
| PRMT1 | 13.0702 | 14.2977 | 13.8194 | 10.7267 | 6.9681 | 7.9953 |
| NDUFB1 | 11.4647 | 11.3549 | 10.9823 | 7.2298 | 8.9176 | 7.8652 |
| PRSS3 | 6.7778 | 6.6156 | 5.6122 | 4.0381 | 4.3812 | 4.2246 |
| ANXA13 | 8.4562 | 9.0276 | 9.5961 | 7.3199 | 6.6528 | 6.6711 |
| RPS14P3 | 12.754 | 13.306 | 12.8153 | 10.6687 | 9.7724 | 8.1203 |
| PRDX1 | 10.1075 | 11.0742 | 10.5858 | 8.5962 | 5.6701 | 7.2069 |
| HNRNPUL1 | 10.9493 | 11.3629 | 10.6094 | 8.9352 | 7.7837 | 7.9296 |
| ID1 | 9.6433 | 9.8069 | 11.0557 | 4.078 | 7.5112 | 6.282 |
| SLC25A11 | 12.262 | 12.6789 | 11.3904 | 9.2069 | 8.692 | 10.2874 |
| TNFRSF10B | 8.3202 | 7.7911 | 8.68 | 4.7971 | 3.4882 | 4.8622 |
| ANXA3 | 9.2352 | 8.1361 | 9.0684 | 5.0505 | 5.1385 | 5.7836 |
| COA1 | 8.8852 | 9.6213 | 9.9783 | 6.4513 | 7.1 | 7.6233 |
| NSL1 | 9.2222 | 8.5947 | 8.8515 | 6.5389 | 3.9251 | 6.3366 |
| CRY1 | 8.0533 | 7.8389 | 8.8556 | 5.4979 | 5.3067 | 5.488 |
| CDKN3 | 11.9004 | 11.434 | 11.8734 | 7.4545 | 7.9377 | 8.6793 |
| APEX1 | 13.7348 | 15 | 14.0462 | 12.393 | 11.2323 | 12.0069 |
| B9D1 | 11.83 | 12.0763 | 11.9587 | 9.8027 | 9.7265 | 9.8755 |
| SPINT2 | 10.2114 | 11.2768 | 11.0265 | 8.3758 | 8.1551 | 8.6273 |
| SERPINB6 | 11.4419 | 11.7332 | 11.7038 | 7.3813 | 7.2291 | 8.7964 |
| EIF4A1 | 13.9494 | 15 | 14.3906 | 11.844 | 11.863 | 9.999 |
| SMARCE1 | 11.6672 | 11.0128 | 11.5652 | 9.4771 | 7.552 | 7.7553 |
| U2SURP | 10.8538 | 10.5268 | 11.0501 | 7.7229 | 8.2864 | 9.3393 |
| CBX5 | 11.4085 | 11.3789 | 11.5185 | 9.5059 | 9.858 | 9.1339 |
| NUP205 | 10.2955 | 10.6101 | 10.7789 | 7.9002 | 8.4982 | 8.3078 |
| CHMP7 | 8.2452 | 9.1627 | 8.7137 | 5.7535 | 6.9781 | 6.5576 |
| LIMCH1 | 14.243 | 14.164 | 15 | 9.1516 | 9.6794 | 11.3703 |
| NBEAL2 | 10.3396 | 9.7061 | 9.107 | 6.8983 | 6.4589 | 7.9466 |
| BBS4 | 8.3403 | 8.4893 | 8.5853 | 6.5738 | 6.5293 | 6.5452 |
| GSAP | 5.4563 | 5.4485 | 6.1753 | 3.2362 | 2.1521 | 1.0287 |
| ZNF23 | 7.1874 | 7.3475 | 7.8826 | 5.5309 | 5.3853 | 4.8161 |
| HNRNPA1 | 12.9634 | 12.6546 | 11.9946 | 10.1707 | 10.8298 | 9.823 |
| SPG7 | 11.643 | 11.3738 | 11.3105 | 9.9434 | 9.3405 | 8.8718 |
| CXCL5 | 8.408 | 7.8511 | 8.9513 | 3.4239 | 0.7879 | 0 |
| SDHC | 12.8323 | 13.3402 | 12.2908 | 10.7698 | 9.6476 | 10.9517 |
| EXOSC8 | 11.5121 | 11.5134 | 11.9015 | 8.9412 | 6.3505 | 7.988 |
| GGCT | 11.0387 | 10.4087 | 10.1788 | 6.5778 | 7.3788 | 8.8144 |
| ATP2B1 | 9.0993 | 7.918 | 9.518 | 5.2431 | 6.3016 | 5.0193 |
| COPZ1 | 10.8542 | 12.0628 | 10.9251 | 8.9361 | 8.1021 | 8.5834 |
| UBE2Z | 9.0829 | 9.3492 | 9.5973 | 7.4496 | 6.3337 | 6.1463 |
| RDH11 | 9.3242 | 8.4735 | 9.3515 | 6.2979 | 6.2578 | 7.1118 |
| PRMT5 | 10.6848 | 11.812 | 11.6791 | 9.6839 | 8.963 | 9.0609 |
| NUCKS1 | 13.171 | 12.893 | 13.3188 | 9.0058 | 10.7104 | 8.3423 |
| POLDIP2 | 10.595 | 10.4798 | 10.6537 | 8.959 | 8.279 | 8.0821 |
| SLTM | 7.9562 | 8.1659 | 7.876 | 6.26 | 5.9308 | 5.9772 |
| CDC27 | 9.9397 | 10.1725 | 9.6098 | 8.038 | 7.7474 | 6.4488 |
| ASNSD1 | 7.9948 | 8.4967 | 9.1345 | 6.4066 | 5.3966 | 4.9523 |
| FKBP3 | 13.0006 | 10.7759 | 12.9284 | 8.4727 | 8.1684 | 8.1648 |
| COA3 | 14.4702 | 13.6304 | 15 | 11.1868 | 11.3738 | 11.3491 |
| AMZ2 | 11.9726 | 11.1854 | 10.1725 | 8.4088 | 7.0575 | 8.66 |
| DHX40 | 11.4325 | 11.7892 | 11.8572 | 8.4136 | 7.5252 | 9.6098 |
| ZWILCH | 9.9979 | 9.5411 | 10.5009 | 8.3929 | 6.1582 | 6.5257 |
| SNF8 | 11.5347 | 11.8001 | 11.7729 | 9.4136 | 7.9803 | 9.1365 |
| SFXN1 | 10.422 | 9.9295 | 10.9711 | 8.4957 | 8.4173 | 8.5185 |
| ACTR6 | 8.5224 | 7.99 | 9.5666 | 6.5112 | 5.7518 | 5.8009 |
| CMC2 | 11.41 | 10.8503 | 12.372 | 9.14 | 7.8085 | 7.6653 |
| GLYR1 | 9.41 | 9.9248 | 9.0362 | 7.0931 | 7.2367 | 6.8312 |
| WDR12 | 9.3055 | 9.4605 | 9.6715 | 6.6606 | 7.1712 | 6.5012 |
| SLC25A15 | 8.0871 | 7.4929 | 7.8695 | 6.3073 | 5.347 | 5.1401 |
| MRPS33 | 11.1628 | 11.0695 | 11.2931 | 7.8808 | 7.4429 | 7.409 |
| MUC13 | 7.6892 | 6.9578 | 7.6007 | 5.2793 | 2.4391 | 4.5514 |
| SCML1 | 7.3267 | 6.6997 | 5.3961 | 2.1464 | 3.1849 | 3.7147 |
| RPL26L1 | 11.5277 | 11.1865 | 12.6254 | 7.7415 | 9.5638 | 8.9444 |
| ACSF2 | 7.6164 | 7.9367 | 7.9807 | 5.8755 | 5.9679 | 6.3308 |
| MED23 | 8.6889 | 9.2514 | 8.8085 | 6.2682 | 6.5371 | 6.9371 |
| NETO2 | 11.8465 | 11.6105 | 13.3265 | 9.1274 | 6.8927 | 9.4479 |
| PLAC8 | 6.3517 | 5.0798 | 6.5493 | 2.0492 | 3.5113 | 0 |
| PLEKHA1 | 9.1459 | 9.247 | 9.5802 | 6.6578 | 7.7881 | 6.7325 |
| SYNJ2BP | 10.5506 | 10.6698 | 10.8133 | 8.4708 | 6.1421 | 7.6342 |
| ZBTB38 | 7.6757 | 7.9107 | 8.4604 | 3.7127 | 4.7929 | 3.3393 |
| TMEM100 | 5.2162 | 6.1017 | 4.6161 | 2.6325 | 2.0795 | 3.4804 |
| TIPIN | 8.7954 | 9.3892 | 9.1778 | 6.582 | 6.4775 | 6.1843 |
| OLA1 | 11.3032 | 11.3064 | 11.0546 | 8.6874 | 7.3074 | 9.517 |
| FA2H | 7.5337 | 7.1195 | 7.3081 | 5.2387 | 2.8305 | 2.6484 |
| ZNF12 | 10.6304 | 9.2173 | 10.3948 | 8.1313 | 6.7367 | 7.6528 |
| PCDH9 | 9.8245 | 9.2903 | 8.1649 | 6.4496 | 3.4612 | 1.7222 |
| DEPDC1 | 8.3966 | 8.3461 | 8.394 | 7.0371 | 5.7815 | 5.5355 |
| PPA2 | 9.5445 | 11.0733 | 10.6339 | 6.8212 | 6.1913 | 6.6645 |
| EMC6 | 12.8282 | 12.6136 | 13.1165 | 10.4142 | 10.3893 | 9.8141 |
| SF3B5 | 11.8898 | 12.3347 | 12.3949 | 10.08 | 9.2841 | 9.656 |
| MIS12 | 10.2556 | 9.6467 | 10.0993 | 7.6774 | 7.0557 | 6.7025 |
| SPDL1 | 8.3724 | 9.5411 | 9.6533 | 6.6076 | 7.2751 | 6.3456 |
| SPATS2L | 7.3751 | 9.9149 | 8.0364 | 5.2095 | 5.2814 | 3.9771 |
| DZIP3 | 8.5306 | 8.7248 | 8.1616 | 6.4718 | 6.5167 | 6.623 |
PTPN11 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: PTPN11
[1] [PTPN11 and the deafness].
[2] Leukaemogenic effects of Ptpn11 activating mutations in the stem cell microenvironment.
[3] The Clinical impact of PTPN11 mutations in adults with acute myeloid leukemia.
[8] Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling.
[9] PTPN11 hypomethylation is associated with gastric cancer progression.
[10] A PTPN11 mutation in a woman with Noonan syndrome and protein-losing enteropathy.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below