CHD1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN2211
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: PHA-793887
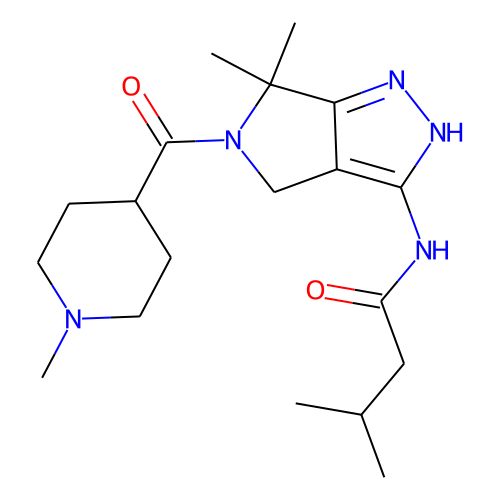
| ID | BRD-K64800655 |
| name | PHA-793887 |
| type | Small molecule |
| molecular formula | C19H31N5O2 |
| molecular weight | 361.5 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | CC(C)CC(=O)Nc1[nH]nc2c1CN(C(=O)C1CCN(C)CC1)C2(C)C |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: CHD1
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CDK1 |
|
|
CDK2 
|
hsa04934 
|
|
CDK4 |
hsa05214 
|
|
CDK5 
|
|
|
CDK7 
|
hsa03420 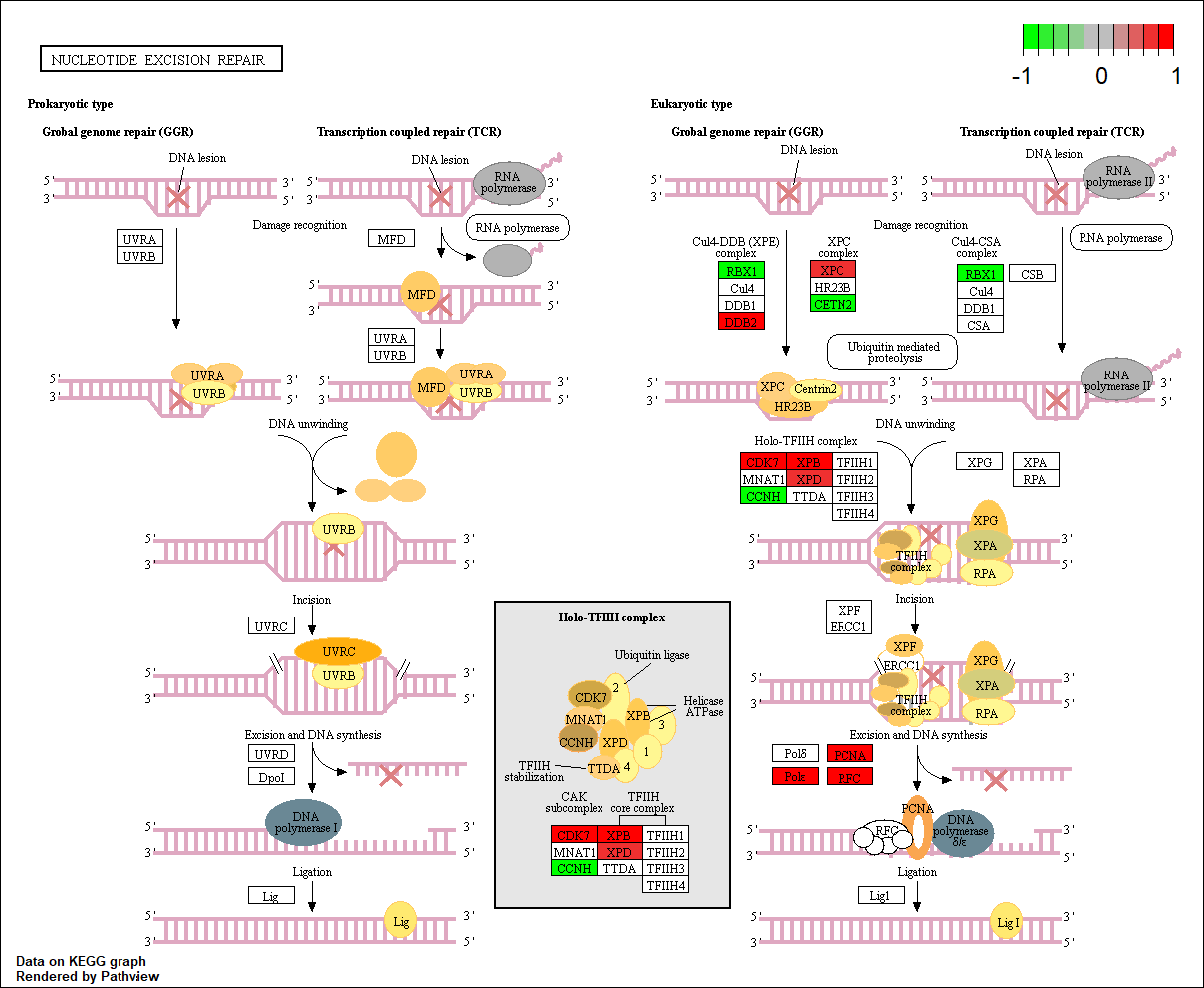
|
|
CDK9 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|---|---|
| ZBTB25 | 5.7915 | 6.196 | 5.8791 | 4.7541 | 4.4051 | 4.6482 | 4.9523 | 4.521 | 5.0543 |
| BRCA1 | 7.2584 | 7.3101 | 6.9423 | 5.728 | 5.8828 | 5.5323 | 5.9125 | 5.3976 | 5.5209 |
| CEBPA | 6.4753 | 6.7784 | 6.0659 | 3.6499 | 3.6692 | 3.2257 | 3.4734 | 3.5003 | 3.3509 |
| CRK | 10.3122 | 10.9164 | 10.0163 | 8.3622 | 8.3898 | 7.7118 | 8.5063 | 6.8353 | 5.8811 |
| CDC20 | 14.3896 | 14.2226 | 14.0075 | 11.7905 | 12.6578 | 12.7566 | 11.9106 | 11.4866 | 10.551 |
| PLK1 | 11.569 | 12.0245 | 12.2935 | 8.2726 | 4.5706 | 7.3935 | 6.4741 | 6.8888 | 8.0528 |
| CDKN1B | 12.4028 | 12.9196 | 13.1055 | 8.7608 | 9.5001 | 10.2578 | 8.8988 | 9.063 | 10.0934 |
| TSKU | 8.2115 | 7.963 | 8.514 | 5.5905 | 6.6908 | 6.0008 | 6.0087 | 4.8843 | 3.6692 |
| WIPF2 | 7.8562 | 7.7821 | 7.9983 | 5.9178 | 6.146 | 6.5924 | 6.3209 | 5.5719 | 5.1722 |
| RRP8 | 6.554 | 6.4324 | 6.5333 | 5.1368 | 5.6412 | 5.1585 | 4.9612 | 5.114 | 5.0164 |
| DNAJB1 | 11.9158 | 11.896 | 12.0156 | 8.6317 | 9.3744 | 8.6349 | 8.1302 | 6.2765 | 5.4486 |
| USP1 | 7.2379 | 7.3516 | 7.6197 | 5.0484 | 5.7664 | 5.7429 | 5.5052 | 4.9679 | 4.8491 |
| KIF2C | 9.5108 | 9.0592 | 9.8733 | 6.8236 | 6.042 | 6.7548 | 6.4372 | 6.1105 | 6.0321 |
| PLA2G15 | 7.2268 | 7.2446 | 7.5625 | 5.5905 | 4.9613 | 5.8843 | 4.8182 | 5.4395 | 4.4912 |
| SLC11A2 | 7.6206 | 7.6611 | 7.6263 | 5.3953 | 6.6136 | 4.5575 | 6.186 | 5.7965 | 5.4357 |
| NET1 | 10.2921 | 9.7334 | 10.3891 | 8.2532 | 8.6198 | 8.3986 | 7.5204 | 6.9854 | 7.1809 |
| ATP1B1 | 9.7766 | 9.6921 | 9.9768 | 8.2119 | 6.6908 | 8.6768 | 7.248 | 7.0789 | 7.5767 |
| GRWD1 | 9.4611 | 9.2265 | 9.181 | 5.2336 | 5.8062 | 5.6691 | 5.4599 | 5.6985 | 3.7296 |
| SMAD3 | 7.814 | 8.0445 | 8.1857 | 5.9817 | 4.6733 | 6.2197 | 6.2038 | 5.3652 | 5.0054 |
| CDCA4 | 7.7664 | 7.8293 | 9.6802 | 5.0218 | 5.4567 | 6.0421 | 5.3068 | 4.9044 | 4.6969 |
| CHMP6 | 6.1077 | 6.4057 | 6.6859 | 4.4465 | 5.3124 | 4.9069 | 4.4361 | 5.3652 | 5.0617 |
| CGRRF1 | 10.0163 | 9.8051 | 10.1891 | 8.3521 | 8.2008 | 8.4716 | 8.7735 | 7.715 | 6.9521 |
| ERBB2 | 9.9166 | 9.4979 | 10.0332 | 6.6045 | 5.0815 | 6.0502 | 7.2614 | 7.2196 | 7.1594 |
| KIF14 | 8.5563 | 8.9416 | 8.8689 | 6.042 | 6.7644 | 3.5705 | 5.6901 | 5.1357 | 5.5572 |
| PARP2 | 9.3098 | 9.5765 | 9.8649 | 6.1817 | 7.3328 | 6.0666 | 5.9221 | 5.2574 | 7.1373 |
| CCNF | 8.8562 | 9.264 | 10.5942 | 4.9156 | 8.2108 | 5.4776 | 4.5245 | 4.6232 | 4.3953 |
| FADD | 9.2753 | 9.6842 | 9.5578 | 7.5715 | 6.8599 | 7.4938 | 7.648 | 6.8723 | 5.9411 |
| TRIP10 | 8.1547 | 8.6932 | 8.5823 | 7.3915 | 6.5361 | 7.6216 | 7.0403 | 6.6895 | 6.6794 |
| ZNF629 | 7.5013 | 7.1166 | 7.744 | 5.9834 | 5.8907 | 5.7989 | 5.2692 | 6.003 | 5.5498 |
| ZC3H4 | 8.239 | 7.9668 | 7.8277 | 5.803 | 6.2485 | 6.7208 | 5.5721 | 6.0379 | 5.1354 |
| CEP55 | 8.7177 | 9.0149 | 8.5667 | 7.5371 | 6.612 | 6.9213 | 7.0375 | 7.2825 | 6.4198 |
| DDX39A | 14.5452 | 14.7834 | 14.0494 | 11.9572 | 10.1533 | 12.29 | 12.3142 | 11.931 | 10.6935 |
| CDC123 | 9.5884 | 10.4397 | 9.7654 | 8.2682 | 8.3426 | 7.4369 | 7.3801 | 8.8414 | 8.3431 |
| TNFAIP2 | 5.5457 | 5.3825 | 6.107 | 3.6986 | 3.5582 | 3.6626 | 4.8272 | 3.9994 | 3.5364 |
| TOB1 | 11.5616 | 11.8343 | 11.2585 | 8.7551 | 8.1847 | 9.9039 | 9.5714 | 9.0264 | 9.4879 |
| SPAG5 | 9.4895 | 9.626 | 10.0637 | 8.1635 | 6.9762 | 8.2875 | 7.287 | 7.7995 | 6.7773 |
| ARF6 | 11.4096 | 11.3128 | 11.0676 | 10.3948 | 10.5359 | 10.034 | 10.0242 | 10.1676 | 10.0066 |
| MED1 | 7.547 | 8.2842 | 7.8071 | 5.9613 | 5.83 | 6.7779 | 5.6725 | 5.823 | 4.778 |
| FSTL3 | 7.1795 | 7.19 | 7.3238 | 5.3914 | 4.3624 | 5.6301 | 5.9153 | 4.8976 | 5.1342 |
| GCLM | 8.2055 | 8.3893 | 8.2203 | 6.2048 | 5.7875 | 6.8654 | 6.2782 | 4.9858 | 5.2667 |
| TRIM16 | 10.1211 | 10.103 | 10.0416 | 7.9152 | 6.622 | 8.5938 | 8.0054 | 7.1651 | 5.8844 |
| EXO1 | 8.1537 | 8.4444 | 8.2258 | 6.5052 | 7.3674 | 6.7334 | 6.8801 | 6.3904 | 7.2567 |
| IDI1 | 11.4427 | 11.3553 | 10.9028 | 9.2323 | 6.8057 | 8.4558 | 8.9822 | 8.9189 | 8.7325 |
| TROAP | 9.2753 | 9.1709 | 9.4151 | 8.0792 | 7.2266 | 7.5532 | 7.4886 | 7.7506 | 8.6208 |
| FEN1 | 12.273 | 12.9367 | 12.5266 | 10.7885 | 9.6655 | 10.0641 | 10.7283 | 10.3719 | 10.1542 |
| MLH3 | 6.4411 | 6.7305 | 6.9733 | 5.1475 | 5.914 | 5.0531 | 5.3505 | 4.9144 | 4.8437 |
| BCL3 | 7.8094 | 6.5054 | 7.5821 | 5.3049 | 5.2458 | 5.9262 | 4.7559 | 5.5563 | 5.1108 |
| URB2 | 7.2294 | 7.3077 | 7.2302 | 5.9707 | 6.2042 | 5.4159 | 5.8786 | 5.6276 | 5.8259 |
| SAC3D1 | 10.4251 | 10.5241 | 9.9079 | 8.8376 | 8.3001 | 8.4596 | 8.6944 | 7.9656 | 6.7385 |
| GEMIN4 | 9.5386 | 9.4448 | 9.1247 | 7.366 | 6.8164 | 7.6537 | 7.0416 | 7.5965 | 6.4004 |
| ALKBH1 | 7.0718 | 7.5458 | 8.0081 | 5.508 | 5.9737 | 5.8751 | 6.4514 | 5.6309 | 5.7596 |
| KIN | 8.5902 | 8.5691 | 8.6706 | 7.2406 | 6.3314 | 7.2563 | 6.5615 | 6.173 | 6.7323 |
| BLM | 6.159 | 6.4016 | 6.5125 | 3.8616 | 3.3016 | 3.5545 | 3.9446 | 3.725 | 4.0845 |
| SLC27A2 | 5.498 | 6.3033 | 6.6042 | 3.1383 | 3.4207 | 3.8638 | 2.8883 | 2.8285 | 0.7277 |
| SIX1 | 8.4408 | 8.2514 | 8.1878 | 6.1654 | 5.6915 | 6.5669 | 5.8959 | 6.3892 | 4.9656 |
| CHRNA5 | 6.3617 | 6.6869 | 6.3555 | 4.7983 | 4.2019 | 4.8297 | 5.0508 | 4.8439 | 5.119 |
| HOXC6 | 6.7349 | 9.398 | 7.4266 | 3.8975 | 3.443 | 3.7068 | 3.2485 | 0 | 0 |
| ARHGAP12 | 6.3948 | 6.6681 | 6.4663 | 4.5602 | 4.0072 | 5.4755 | 4.6434 | 5.2729 | 5.074 |
| CENPF | 6.6482 | 7.2035 | 6.6425 | 5.8184 | 5.3988 | 5.1021 | 5.4735 | 5.3354 | 5.8281 |
| EHD1 | 7.7881 | 8.0743 | 8.2353 | 5.5564 | 5.7522 | 6.4361 | 5.6532 | 6.718 | 5.2527 |
| SEPHS1 | 9.4994 | 9.9735 | 9.9693 | 8.1029 | 5.7316 | 7.2566 | 6.2682 | 7.5058 | 7.2757 |
| AATF | 9.1709 | 9.471 | 8.8432 | 7.6008 | 7.9031 | 7.8033 | 7.62 | 7.9946 | 7.7728 |
| CDR2 | 5.9931 | 5.7025 | 6.701 | 4.6347 | 4.9993 | 4.8312 | 4.6965 | 3.9357 | 4.1518 |
| ING1 | 6.4378 | 6.7351 | 6.3818 | 4.6548 | 5.5778 | 5.0659 | 5.0352 | 4.7549 | 4.8456 |
| FRAT2 | 7.9743 | 8.7461 | 7.5068 | 5.2554 | 5.3888 | 6.1776 | 5.8894 | 4.3595 | 5.0998 |
| TAF5 | 8.1796 | 7.8516 | 7.7375 | 6.8255 | 6.8918 | 6.5506 | 6.3831 | 5.8593 | 6.4373 |
| GEMIN2 | 8.3539 | 8.736 | 8.4842 | 7.374 | 7.0613 | 6.9228 | 7.7365 | 7.045 | 6.4055 |
| HSP90AA1 | 13.6869 | 14.8821 | 15 | 12.3984 | 12.1965 | 12.5307 | 12.7363 | 11.8479 | 13.0683 |
| ERAL1 | 10.032 | 9.86 | 10.2754 | 8.8684 | 8.3289 | 9.3116 | 8.797 | 8.751 | 8.5117 |
| CARM1 | 12.1858 | 11.6948 | 12.5692 | 10.909 | 10.7637 | 10.8159 | 10.9516 | 10.0641 | 10.0652 |
| ARHGAP19 | 7.3919 | 7.3737 | 6.9287 | 5.6784 | 5.3012 | 5.8215 | 6.1939 | 4.8101 | 4.5059 |
| CKAP5 | 9.4219 | 9.5353 | 9.7547 | 8.3394 | 7.8574 | 8.787 | 8.0688 | 8.3398 | 7.5571 |
| MFSD5 | 9.8462 | 9.8415 | 9.6025 | 8.7208 | 7.507 | 7.8556 | 8.4657 | 7.2777 | 7.4994 |
| TEX30 | 8.5805 | 9.0385 | 8.8909 | 7.2186 | 7.1636 | 6.4825 | 7.2507 | 6.3608 | 5.8892 |
| ZBTB1 | 8.0117 | 7.6512 | 8.0125 | 5.9918 | 5.2025 | 6.7759 | 5.9645 | 5.7006 | 5.3134 |
| PCGF2 | 8.7293 | 9.3759 | 9.1697 | 6.9153 | 7.4059 | 7.5333 | 6.5694 | 6.1725 | 4.9747 |
| HNRNPA1 | 12.3114 | 12.3698 | 12.3883 | 10.9855 | 10.4155 | 11.4726 | 10.2897 | 10.8633 | 10.8813 |
| MPRIP | 8.4914 | 8.1068 | 8.3842 | 6.0756 | 6.4449 | 7.2847 | 6.4326 | 7.1358 | 5.6983 |
| WAC | 8.0851 | 8.1983 | 8.0908 | 6.5669 | 5.3403 | 6.3456 | 5.7266 | 5.5052 | 5.0575 |
| PRC1 | 8.881 | 9.4715 | 9.8307 | 8.083 | 6.9876 | 7.68 | 6.8494 | 6.808 | 7.2061 |
| ASF1B | 10.5784 | 10.8999 | 10.6468 | 9.8836 | 9.7331 | 9.1673 | 9.3802 | 9.5232 | 9.0704 |
| CENPT | 8.8688 | 8.6362 | 8.6529 | 7.7015 | 7.1853 | 7.9677 | 6.9867 | 7.5528 | 7.3713 |
| SAV1 | 7.9598 | 8.0625 | 8.0966 | 6.5925 | 6.391 | 6.324 | 6.493 | 5.8504 | 4.9073 |
| METTL17 | 11.9741 | 11.7516 | 12.0838 | 10.6278 | 10.876 | 11.0598 | 10.8704 | 10.6489 | 10.9897 |
| S100PBP | 7.2221 | 7.3891 | 6.7759 | 5.7094 | 6.1507 | 5.4011 | 5.6072 | 6.0329 | 5.1998 |
| TTC4 | 8.7045 | 8.719 | 8.5394 | 7.8799 | 7.3277 | 7.7234 | 7.7545 | 7.6825 | 7.3354 |
| FAM222B | 6.7185 | 6.8843 | 7.1007 | 5.1254 | 5.0621 | 5.0646 | 5.1489 | 4.0394 | 4.1666 |
| PPP1R13L | 7.2165 | 7.7347 | 8.0447 | 6.3945 | 6.5022 | 5.9067 | 6.0535 | 5.1192 | 5.5128 |
| RNF121 | 6.8409 | 6.7509 | 6.6377 | 5.4184 | 5.7232 | 5.6623 | 5.7092 | 5.6534 | 4.878 |
| RHOF | 7.7345 | 7.5221 | 6.8534 | 5.7607 | 4.3427 | 4.4531 | 5.8024 | 3.2697 | 3.7851 |
| HSPA14 | 9.1016 | 9.3098 | 9.9506 | 7.1143 | 7.5849 | 6.532 | 5.9666 | 6.2067 | 5.5301 |
| COA7 | 8.5013 | 8.3445 | 8.3515 | 7.1612 | 7.0489 | 7.3398 | 6.9585 | 6.9449 | 7.7286 |
| CLCF1 | 6.57 | 6.6728 | 7.0991 | 5.0182 | 5.8221 | 5.6127 | 5.4334 | 5.0209 | 4.5288 |
| CHST11 | 6.4987 | 5.8029 | 5.6175 | 4.5989 | 4.2792 | 4.5543 | 4.4847 | 4.21 | 4.1054 |
| BANP | 8.3502 | 8.8229 | 7.8689 | 6.2301 | 6.4732 | 6.7549 | 7.1007 | 6.6855 | 7.2011 |
| DEPDC1 | 7.9917 | 7.7661 | 7.5187 | 6.0913 | 5.7848 | 6.2737 | 6.5886 | 6.8034 | 6.3709 |
| CENPJ | 4.9322 | 4.6826 | 5.3651 | 3.7053 | 3.7328 | 3.9379 | 3.728 | 3.9936 | 3.8245 |
| KIF18A | 8.0758 | 8.4406 | 7.692 | 6.6223 | 6.332 | 6.8872 | 7.0006 | 6.3768 | 7.1202 |
| CDCA8 | 11.1768 | 11.2186 | 11.5149 | 10.0074 | 8.4461 | 9.0754 | 9.4177 | 9.1795 | 9.7618 |
| HSPB8 | 11.9929 | 12.5954 | 12.6327 | 10.2809 | 8.4254 | 10.0516 | 10.2442 | 9.3866 | 7.9394 |
| IMP3 | 9.7941 | 9.9882 | 9.8033 | 7.9511 | 6.0931 | 7.5364 | 8.3521 | 6.6328 | 6.7312 |
| INTS5 | 8.1727 | 8.3575 | 8.2739 | 7.0396 | 6.7198 | 7.0233 | 7.0397 | 6.7249 | 6.4636 |
| GPSM2 | 10.4753 | 10.4196 | 9.9481 | 9.1762 | 8.6485 | 8.5502 | 9.3194 | 8.7018 | 8.8813 |
| RACGAP1 | 10.7735 | 10.3563 | 10.4934 | 9.1397 | 9.3246 | 9.6742 | 9.0441 | 8.7704 | 8.5127 |
| CENPN | 8.3371 | 8.4201 | 7.7402 | 6.7607 | 6.2774 | 6.3541 | 6.654 | 5.9716 | 5.5696 |
| RASSF4 | 7.6647 | 7.4844 | 6.6011 | 6.1074 | 5.9253 | 5.5847 | 5.361 | 5.3849 | 5.0828 |
| BRF1 | 7.7106 | 7.6326 | 7.7815 | 6.4801 | 6.4507 | 6.4194 | 6.1874 | 6.4321 | 6.5303 |
CHD1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: CHD1
[1] SPOP-Mutated/CHD1-Deleted Lethal Prostate Cancer and Abiraterone Sensitivity.
[3] Nucleosome-Chd1 structure and implications for chromatin remodelling.
[4] Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer.
[9] The chromatin remodeler Chd1 regulates cohesin in budding yeast and humans.
[10] CHD1 and SPOP synergistically protect prostate epithelial cells from DNA damage.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below