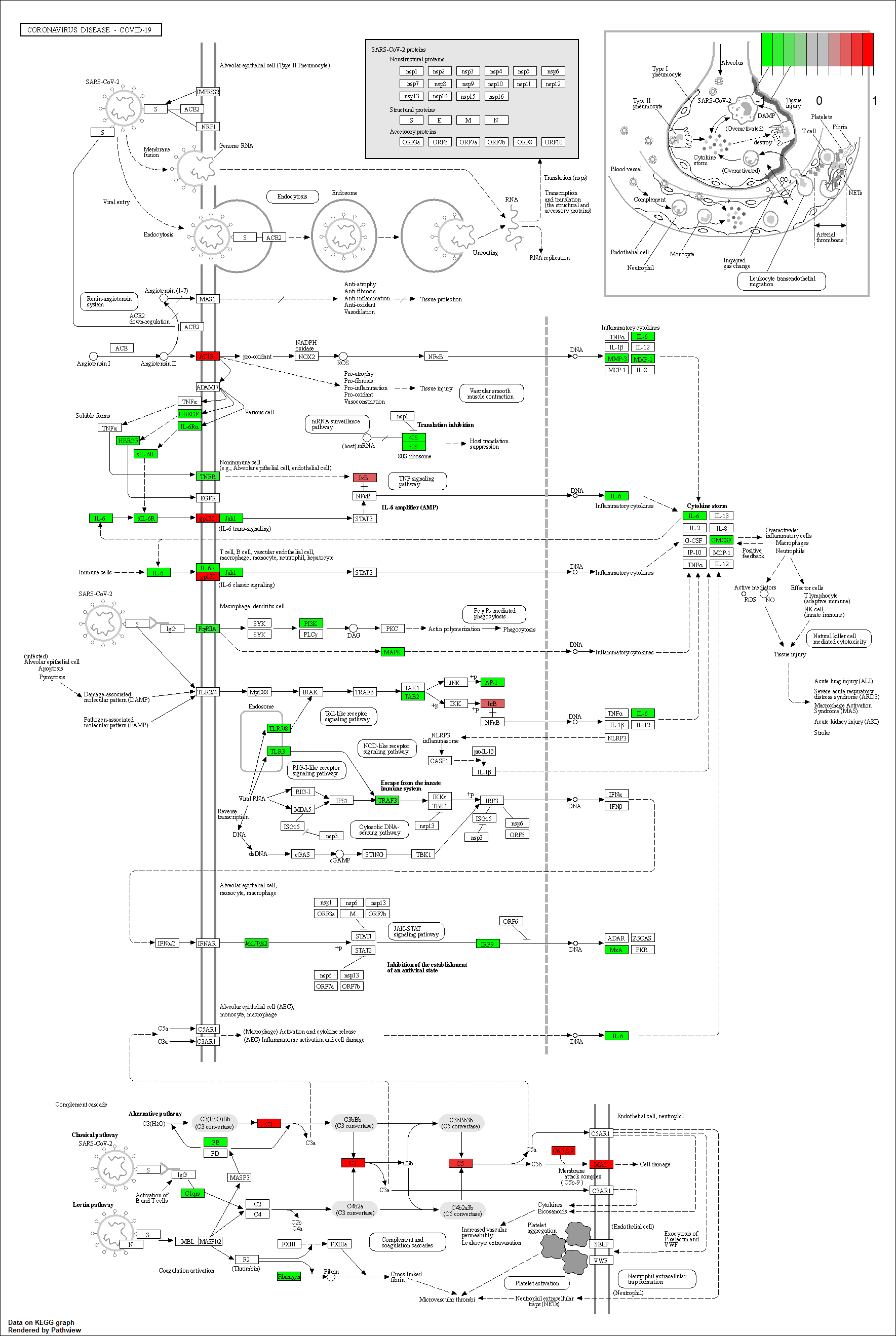
FOS transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN2475
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: SB-218078
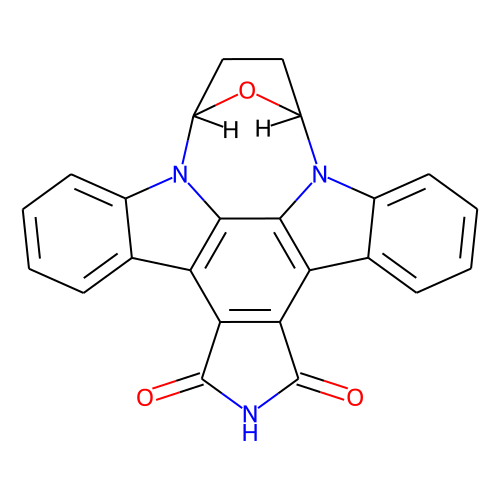
| ID | BRD-A06352508 |
| name | SB-218078 |
| type | Small molecule |
| molecular formula | C24H15N3O3 |
| molecular weight | 393.4 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | "O=C1NC(=O)c2c1c1c3ccccc3n3[C@H]4CC[C@@H](O4)n4c5ccccc5c2c4c13 |&1:15,&2:18,r|" |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: FOS
Transcription regulatory factors corresponding to this network
ARTICLE: SB-218078
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CHEK1 
|
hsa04110 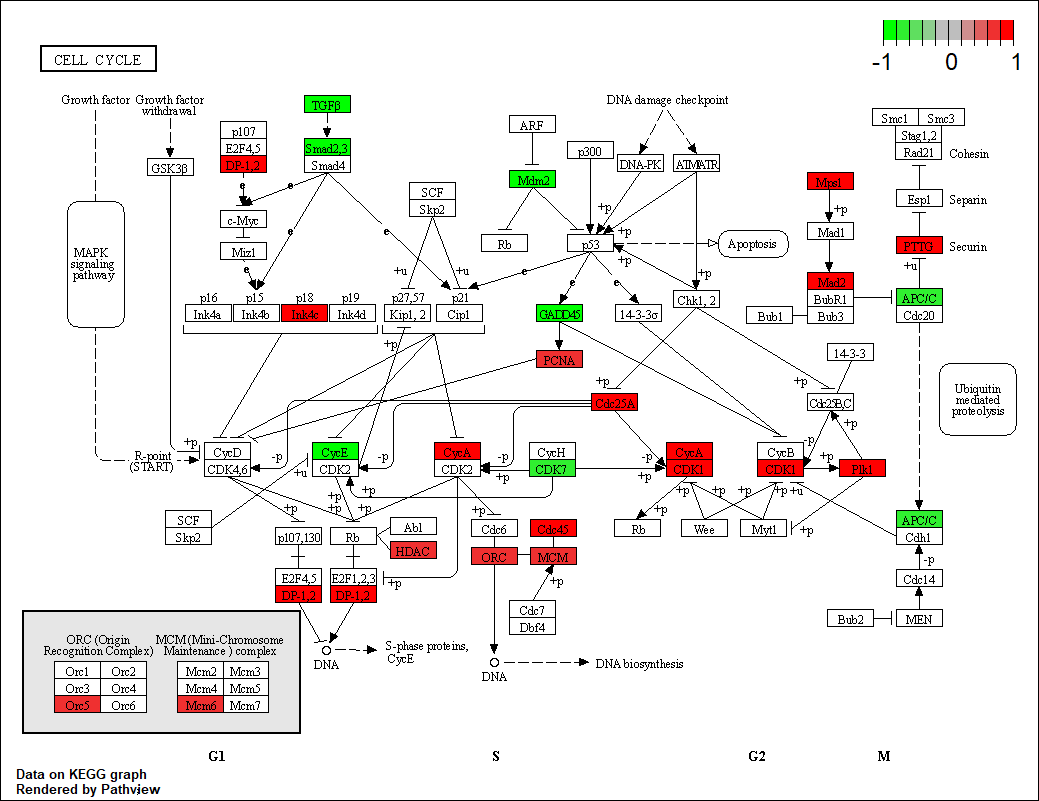
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|
| IL1A | 3.7726 | 4.4593 | 4.5753 | 5.0935 | 6.0589 | 5.5829 | 5.664 |
| IQCB1 | 7.5303 | 7.3658 | 7.3045 | 8.044 | 8.904 | 8.1383 | 8.6269 |
| CLIC5 | 1.6927 | 1.794 | 3.904 | 4.955 | 4.4625 | 3.7947 | 7.0231 |
| PDLIM5 | 6.8861 | 6.7576 | 6.9604 | 8.761 | 7.6977 | 8.6318 | 9.8411 |
| INSR | 7.9949 | 7.193 | 8.6027 | 9.0825 | 9.7601 | 8.7246 | 9.9096 |
| DOK5 | 5.3036 | 5.3373 | 6.1209 | 7.2161 | 6.924 | 7.0154 | 9.1486 |
| NHLH2 | 4.9339 | 5.1065 | 5.4945 | 5.7191 | 6.3713 | 5.8405 | 6.4594 |
| ORAI2 | 5.6897 | 5.9659 | 6.2064 | 7.1328 | 6.7897 | 6.7144 | 6.6896 |
| IL20RA | 5.8848 | 5.1434 | 5.8541 | 6.8623 | 7.489 | 6.7512 | 8.2186 |
| FZD10 | 4.7501 | 4.4781 | 5.5625 | 6.3582 | 6.0127 | 6.168 | 8.1052 |
| TMPRSS3 | 6.6173 | 5.3879 | 7.0841 | 7.7724 | 7.6181 | 7.9624 | 7.354 |
| MUC16 | 5.2947 | 4.4722 | 4.7468 | 7.3915 | 7.1457 | 5.9852 | 5.6163 |
| RALGPS2 | 5.1744 | 5.6309 | 5.5871 | 6.3809 | 6.5009 | 6.4169 | 6.2376 |
| CBLC | 6.5311 | 6.1361 | 6.7732 | 7.7958 | 7.7546 | 7.239 | 8.4393 |
| BCAM | 8.2871 | 8.2743 | 9.2057 | 10.0366 | 9.4802 | 9.4363 | 10.5926 |
| ADAP1 | 7.4303 | 6.8192 | 7.4607 | 8.208 | 9.5027 | 7.9107 | 8.5456 |
| SQSTM1 | 12.7794 | 12.4992 | 12.5762 | 13.6595 | 13.8267 | 14.1586 | 13.6834 |
| EGR1 | 4.8983 | 4.7778 | 4.07 | 7.2805 | 8.3555 | 7.5666 | 7.2961 |
| HMOX1 | 6.5538 | 6.3462 | 6.0416 | 9.7339 | 9.1708 | 9.8579 | 9.1607 |
| NFKBIA | 9.3372 | 9.7063 | 10.0877 | 11.5133 | 12.0942 | 12.7166 | 11.5634 |
| IGFBP3 | 5.4346 | 4.7181 | 4.2436 | 6.543 | 6.7776 | 6.668 | 5.2432 |
| DUSP4 | 5.8619 | 5.8142 | 5.13 | 7.1357 | 7.3081 | 7.7846 | 6.8913 |
| GADD45A | 7.7403 | 6.939 | 6.9652 | 10.4693 | 10.1952 | 10.1011 | 10.0116 |
| JUN | 6.6876 | 6.4457 | 7.1723 | 8.2612 | 7.614 | 8.4154 | 9.6203 |
| ZFP36 | 9.3868 | 9.3816 | 9.9508 | 11.3835 | 12.1895 | 12.1403 | 11.7833 |
| NFIL3 | 6.6312 | 6.4553 | 5.8066 | 7.2305 | 7.4432 | 7.2579 | 6.8697 |
| TRIB1 | 7.2822 | 6.6997 | 6.0027 | 9.0455 | 8.6896 | 8.7959 | 9.5185 |
| TIPARP | 8.7424 | 8.5653 | 8.0896 | 9.6883 | 9.8899 | 10.1449 | 9.6203 |
| DDB2 | 9.8411 | 10.001 | 9.6945 | 11.1823 | 11.1898 | 11.0564 | 10.1627 |
| IER3 | 11.167 | 11.3461 | 11.5705 | 12.877 | 12.9936 | 13.0242 | 12.1665 |
| ATP1B1 | 9.8509 | 9.8428 | 9.4771 | 11.693 | 11.7524 | 11.8279 | 11.0564 |
| JMJD6 | 11.4923 | 11.4445 | 11.4727 | 12.4264 | 12.6929 | 12.3822 | 11.8211 |
| SLC35F2 | 7.4618 | 7.58 | 6.8151 | 8.4613 | 8.458 | 8.7348 | 7.753 |
| SMAD3 | 7.1862 | 6.7973 | 7.2319 | 8.3972 | 9.1322 | 10.6238 | 9.8997 |
| CHAC1 | 7.3348 | 6.7695 | 8.8823 | 12.6549 | 12.4755 | 8.7348 | 12.4647 |
| FOS | 4.041 | 4.5245 | 4.0493 | 6.7167 | 5.6536 | 6.9172 | 5.9671 |
| RELB | 5.3195 | 5.6287 | 5.2801 | 8.0558 | 8.6331 | 9.2808 | 7.7042 |
| ANKRD10 | 6.7022 | 6.6893 | 6.2624 | 7.7694 | 8.2146 | 8.3442 | 8.6135 |
| EDEM1 | 8.2142 | 7.9549 | 8.5454 | 9.2006 | 9.8558 | 9.5714 | 9.9557 |
| EPHA2 | 7.276 | 5.4324 | 7.8883 | 9.0053 | 7.8991 | 9.5233 | 9.7654 |
| CTDNEP1 | 10.8463 | 10.1599 | 11.0107 | 11.486 | 11.7122 | 11.5805 | 11.4061 |
| NCL | 10.7199 | 10.6493 | 11.2982 | 11.5075 | 12.4205 | 11.9903 | 12.3392 |
| CTSA | 10.8428 | 9.9189 | 10.2957 | 11.6868 | 11.8953 | 11.4908 | 11.0885 |
| ACADVL | 13.0653 | 12.5203 | 12.9933 | 14.8563 | 14.9166 | 13.8934 | 14.697 |
| ZYX | 10.9453 | 10.8514 | 10.7476 | 12.0059 | 11.9643 | 12.3519 | 12.7659 |
| VCL | 10.1378 | 10.746 | 10.8573 | 11.4904 | 13.0494 | 11.7656 | 11.9033 |
| ANXA4 | 9.9846 | 9.7374 | 9.5783 | 10.7095 | 10.6131 | 10.9341 | 10.9913 |
| ETS2 | 5.0073 | 4.563 | 5.6586 | 6.9671 | 6.8543 | 7.3964 | 7.6494 |
| JUNB | 7.0817 | 6.7727 | 7.1098 | 9.5569 | 9.3695 | 9.5268 | 9.0159 |
| TGFBI | 3.5266 | 3.5886 | 6.2364 | 6.6274 | 7.3366 | 8.2477 | 8.6844 |
| IFNGR2 | 7.7657 | 7.4748 | 7.8463 | 8.635 | 9.2808 | 8.6521 | 9.7833 |
| CD55 | 6.9472 | 6.0149 | 6.3904 | 8.7007 | 8.7661 | 7.4693 | 7.728 |
| PPP1R15A | 5.7804 | 5.4862 | 5.2645 | 7.7916 | 7.0017 | 7.5383 | 5.7629 |
| SDC4 | 10.0992 | 8.8268 | 10.2206 | 12.4851 | 12.8684 | 14.9422 | 13.5615 |
| IER2 | 9.429 | 8.9178 | 9.604 | 11.441 | 11.2129 | 11.0209 | 11.0464 |
| UBE2B | 8.6299 | 8.5618 | 8.4319 | 9.1862 | 9.67 | 9.6497 | 9.8881 |
| NR4A1 | 5.3796 | 6.1872 | 6.1243 | 8.347 | 7.4822 | 8.4814 | 8.5542 |
| SEC24D | 6.4292 | 6.2988 | 6.2312 | 7.3492 | 7.0308 | 7.5725 | 7.6841 |
| KLF10 | 4.8721 | 4.6868 | 5.4693 | 6.644 | 8.4449 | 6.7344 | 7.698 |
| AHR | 9.2484 | 8.8864 | 10.0023 | 10.3724 | 10.1635 | 10.688 | 10.2887 |
| SPINT1 | 7.2902 | 7.2218 | 8.4355 | 9.4071 | 8.516 | 8.79 | 10.25 |
| MYO1E | 6.2119 | 6.4667 | 6.9599 | 7.4594 | 7.6787 | 7.8755 | 8.2505 |
| CNOT3 | 8.7176 | 7.793 | 9.5146 | 10.0518 | 9.7887 | 9.9988 | 10.5091 |
| OFD1 | 7.4266 | 7.907 | 7.4222 | 8.8697 | 8.8207 | 8.0781 | 8.4352 |
| TRMT1 | 9.385 | 9.0839 | 9.8456 | 10.764 | 10.4821 | 10.194 | 10.4028 |
| VDR | 6.7066 | 6.7949 | 6.98 | 7.4519 | 7.9805 | 7.4721 | 7.9355 |
| NPIPA1 | 9.6574 | 8.8981 | 9.0995 | 11.1099 | 10.6168 | 10.6222 | 9.5183 |
| NR4A2 | 2.2638 | 4.3721 | 4.3932 | 6.7948 | 5.9636 | 6.0766 | 4.4538 |
| BCL3 | 8.4818 | 7.9123 | 7.937 | 8.8125 | 9.543 | 9.0564 | 8.7291 |
| MAFG | 7.8082 | 7.7036 | 7.6104 | 8.5646 | 8.5096 | 8.6047 | 8.6569 |
| AREG | 7.4557 | 6.7857 | 7.4872 | 10.7719 | 12.0557 | 9.2167 | 9.7172 |
| EGR2 | 4.7086 | 5.9607 | 4.2183 | 6.5798 | 7.1317 | 5.9618 | 7.9703 |
| LIF | 7.1238 | 6.7088 | 7.1339 | 8.0991 | 9.4018 | 9.2065 | 7.4451 |
| CCL20 | 3.2792 | 1.6216 | 0.5374 | 5.3858 | 5.9279 | 5.2115 | 7.9968 |
| TUFT1 | 6.7696 | 6.1416 | 6.3767 | 7.7079 | 8.3974 | 7.7079 | 6.7545 |
| ADORA2B | 8.3532 | 7.4781 | 9.3559 | 10.3609 | 10.8123 | 9.4004 | 10.4562 |
| EGR3 | 6.3421 | 5.6896 | 5.7845 | 7.8913 | 9.1308 | 8.2469 | 6.6519 |
| CPNE1 | 10.2074 | 10.1628 | 9.7575 | 10.897 | 10.7164 | 10.7222 | 10.7719 |
| IL11 | 5.5334 | 5.8176 | 5.8465 | 6.6713 | 6.3851 | 7.07 | 6.4448 |
| CAPN2 | 7.0795 | 7.0061 | 8.5099 | 9.9478 | 9.2793 | 9.1119 | 10.6493 |
| PLXNB2 | 9.045 | 9.6023 | 10.6554 | 12.0636 | 12.4205 | 10.6143 | 12.4633 |
| KLF6 | 6.6753 | 6.5775 | 6.2158 | 8.9966 | 7.8771 | 8.5899 | 8.6301 |
| DDAH1 | 11.1717 | 9.6551 | 11.3221 | 12.2164 | 12.6714 | 12.0573 | 12.607 |
| WBP2 | 9.3135 | 8.3735 | 9.2149 | 10.3659 | 10.0451 | 9.753 | 10.3337 |
| RABGGTB | 10.8974 | 10.5651 | 11.0709 | 11.9 | 12.4843 | 11.7752 | 12.2211 |
| KLF5 | 8.3386 | 6.9074 | 7.704 | 8.9074 | 10.9404 | 10.0605 | 9.9505 |
| DUSP5 | 4.9404 | 6.0022 | 4.3918 | 6.2081 | 7.5301 | 7.8363 | 7.1245 |
| OSBPL3 | 5.3522 | 3.0899 | 4.5928 | 6.8801 | 6.7927 | 5.6314 | 5.515 |
| PLA2G4C | 6.8626 | 6.8857 | 8.0411 | 9.4449 | 7.9452 | 9.2324 | 9.6932 |
| PHLDA2 | 10.4217 | 10.7006 | 11.482 | 13.2691 | 12.0868 | 11.5522 | 12.0779 |
| GRINA | 8.9457 | 9.1679 | 9.5806 | 9.989 | 10.3981 | 10.3883 | 10.4187 |
| RHOB | 10.2746 | 9.9917 | 9.6047 | 12.3138 | 11.3333 | 11.3047 | 10.647 |
| SLC38A10 | 8.222 | 6.6081 | 8.3122 | 9.3112 | 9.184 | 8.6211 | 9.4333 |
| ARHGEF18 | 7.392 | 7.3304 | 8.0355 | 9.0979 | 8.014 | 9.5047 | 9.2055 |
| SNAI2 | 5.5063 | 4.3619 | 5.1514 | 6.7374 | 6.5464 | 7.3528 | 10.2949 |
| MAPK8IP3 | 7.0656 | 7.2259 | 7.4965 | 7.7657 | 8.602 | 8.0317 | 8.2323 |
| NPAS2 | 7.7483 | 7.3563 | 7.5606 | 8.6016 | 9.4277 | 9.0483 | 9.0353 |
| ANXA2 | 12.4014 | 13.1824 | 13.8498 | 14.5802 | 14.163 | 13.93 | 14.697 |
| F2RL1 | 5.9502 | 4.5606 | 6.0318 | 7.047 | 7.5864 | 6.9919 | 7.15 |
| RBM8A | 9.1054 | 9.3934 | 9.0517 | 10.3892 | 10.9522 | 9.5265 | 10.6888 |
| DHRS2 | 5.2801 | 4.5894 | 3.2586 | 6.2819 | 6.7515 | 7.0464 | 5.1185 |
| SLC12A7 | 7.018 | 5.8425 | 7.7316 | 8.3201 | 9.1385 | 7.9706 | 8.0893 |
| TVP23B | 8.2647 | 7.7482 | 7.9761 | 9.2542 | 9.0577 | 9.3976 | 9.5646 |
| IER5 | 9.2555 | 8.8006 | 11.3739 | 12.4464 | 11.8009 | 12.9813 | 14.2476 |
| FNDC3B | 6.9645 | 6.0774 | 6.4488 | 7.1184 | 7.8568 | 7.8686 | 7.7778 |
| CDK5RAP3 | 9.3984 | 10.01 | 10.1728 | 10.8893 | 11.7795 | 10.934 | 10.5149 |
| ZC3H12A | 5.2857 | 5.6943 | 5.7753 | 6.278 | 6.944 | 6.806 | 7.2815 |
| PILRB | 6.2209 | 6.4549 | 6.4258 | 7.3595 | 7.9396 | 7.5827 | 6.6955 |
| APOLD1 | 5.7911 | 6.009 | 5.5717 | 7.7799 | 6.7281 | 6.6763 | 7.9757 |
| GDF15 | 12.9097 | 11.6107 | 13.5245 | 15 | 15 | 14.9422 | 13.9284 |
| SLC38A7 | 6.5264 | 6.3721 | 7.1856 | 7.4058 | 7.2988 | 7.9161 | 7.7599 |
| PPP1R10 | 7.0342 | 6.8269 | 7.6539 | 8.4081 | 8.6465 | 8.2225 | 7.6173 |
| LAMP3 | 1.4237 | 1.1195 | 1.9963 | 4.4295 | 4.2861 | 2.3286 | 3.5184 |
| HDAC9 | 4.9079 | 4.2534 | 4.9685 | 5.1104 | 7.0633 | 5.9607 | 7.0329 |
| DSC3 | 4.596 | 5.3668 | 6.0171 | 6.3189 | 6.5908 | 6.2617 | 6.8414 |
| SELE | 5.0232 | 4.2653 | 5.7164 | 6.5486 | 6.9666 | 5.9759 | 7.6472 |
| KRT75 | 4.8446 | 5.0041 | 5.3827 | 5.909 | 6.367 | 5.8197 | 5.6808 |
| EGR4 | 3.6595 | 3.5602 | 3.7962 | 4.4397 | 4.3071 | 5.2253 | 4.554 |
FOS transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa05171