SMARCA4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN3049
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: spiperone
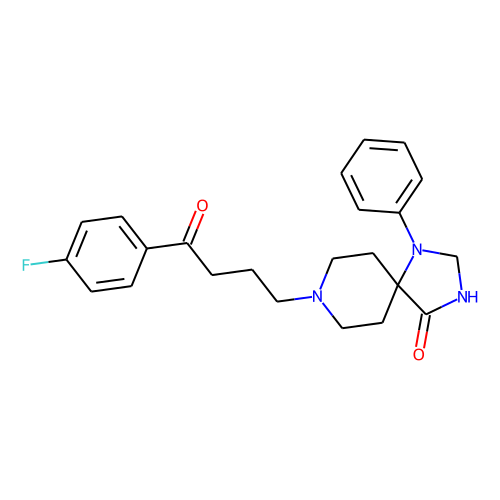
| ID | BRD-K55468218 |
| name | spiperone |
| type | Small molecule |
| molecular formula | C23H26FN3O2 |
| molecular weight | 395.5 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | Fc1ccc(cc1)C(=O)CCCN1CCC2(CC1)N(CNC2=O)c1ccccc1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: HEPG2
Cell lines used for experiments
TRANSCRIPTION FACTOR: SMARCA4
Transcription regulatory factors corresponding to this network
ARTICLE: spiperone
[1] Levodopa inhibits the development of lens-induced myopia in chicks.
[3] Spiperone: Tritium labelling at high specific activity.
[4] Structure of the dopamine D(2) receptor in complex with the antipsychotic drug spiperone.
[5] Distinct Dopamine D₂ Receptor Antagonists Differentially Impact D₂ Receptor Oligomerization.
[7] Ketanserin and spiperone as templates for novel serotonin 5-HT(2A) antagonists.
[9] 125I-Spiperone: a novel ligand for D2 dopamine receptors.
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
ADRA1A 
|
hsa04152 
|
|
ADRA1B |
hsa04970 
|
|
ADRA1D |
hsa04970 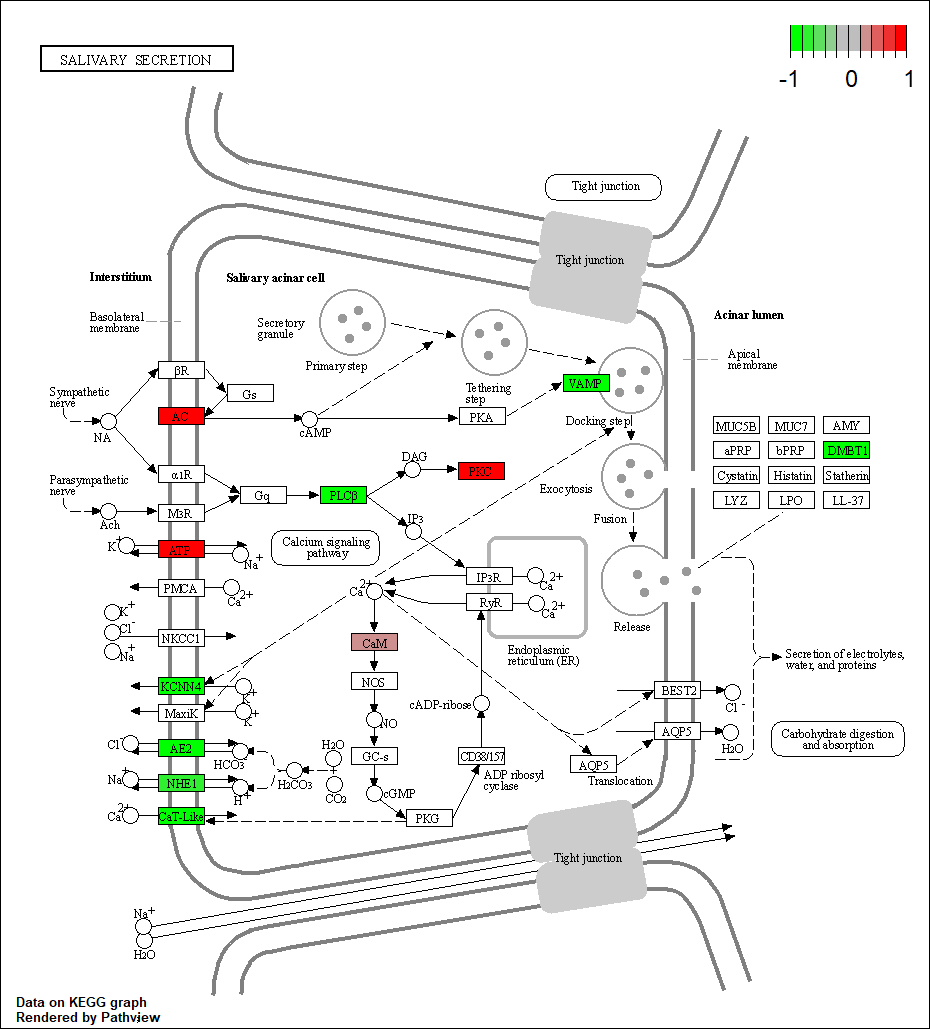
|
|
DRD1 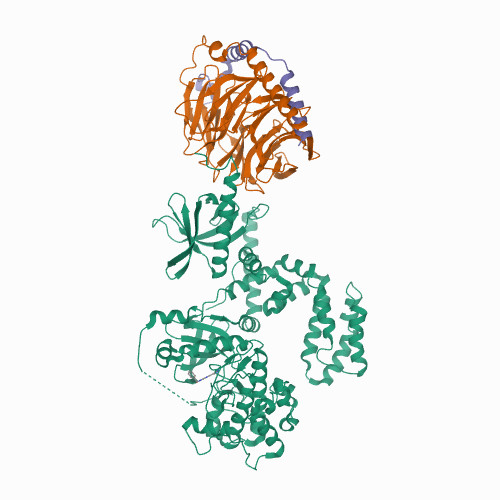
|
hsa04080 
|
|
DRD2 |
hsa05012 
|
|
DRD3 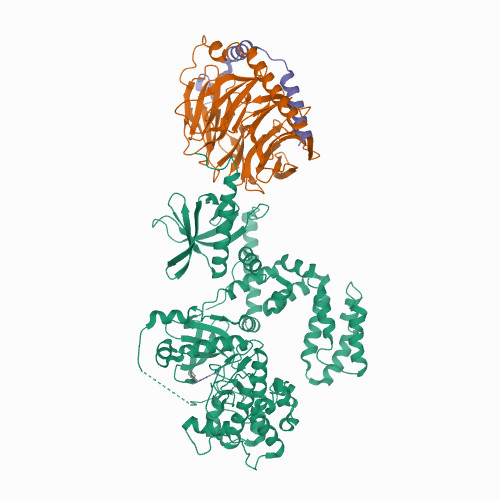
|
|
|
DRD4 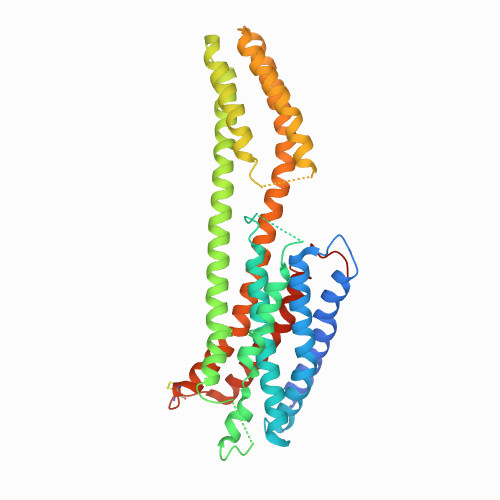
|
|
|
DRD5 |
hsa04728 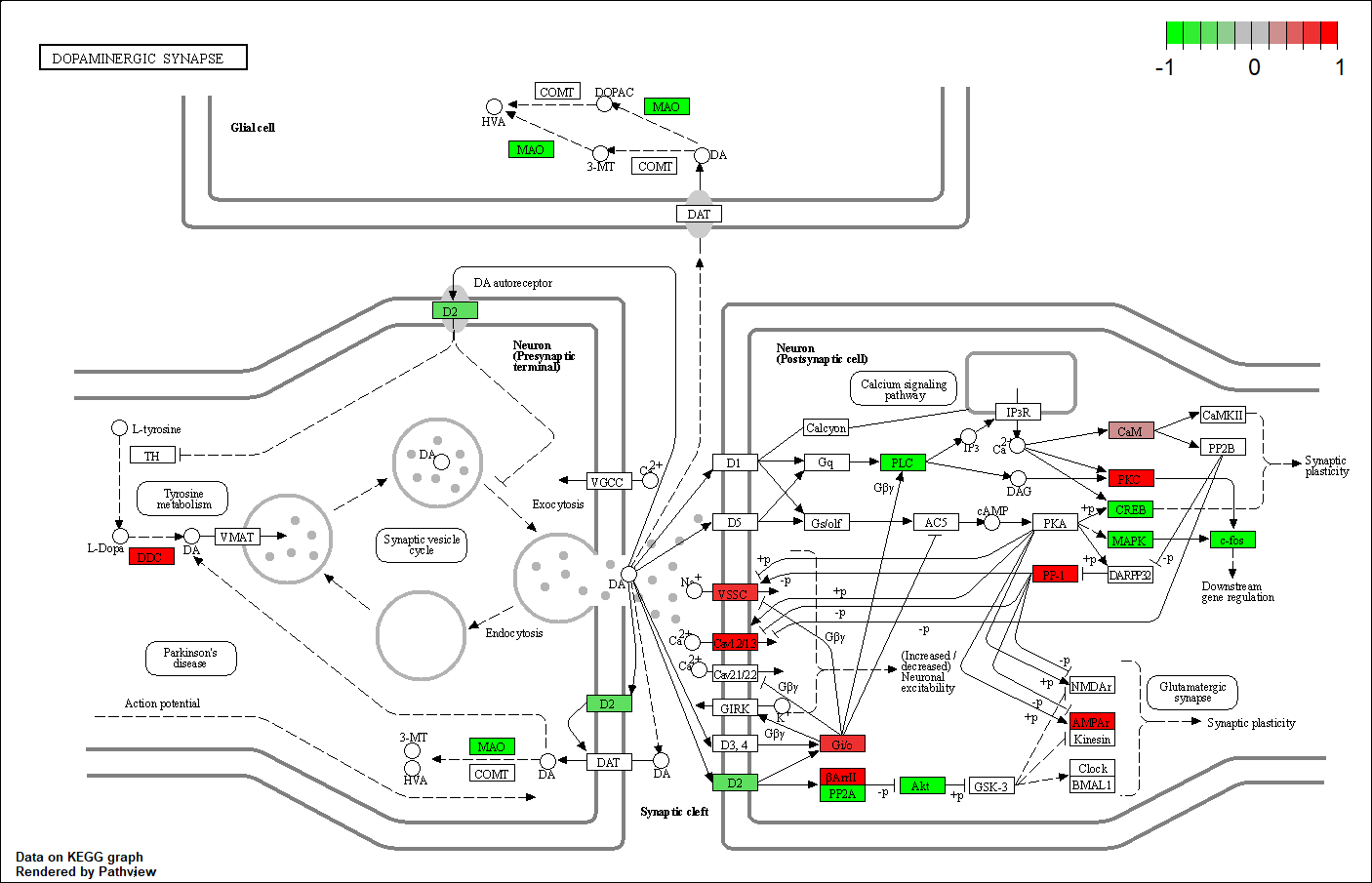
|
|
HTR1A |
hsa04080 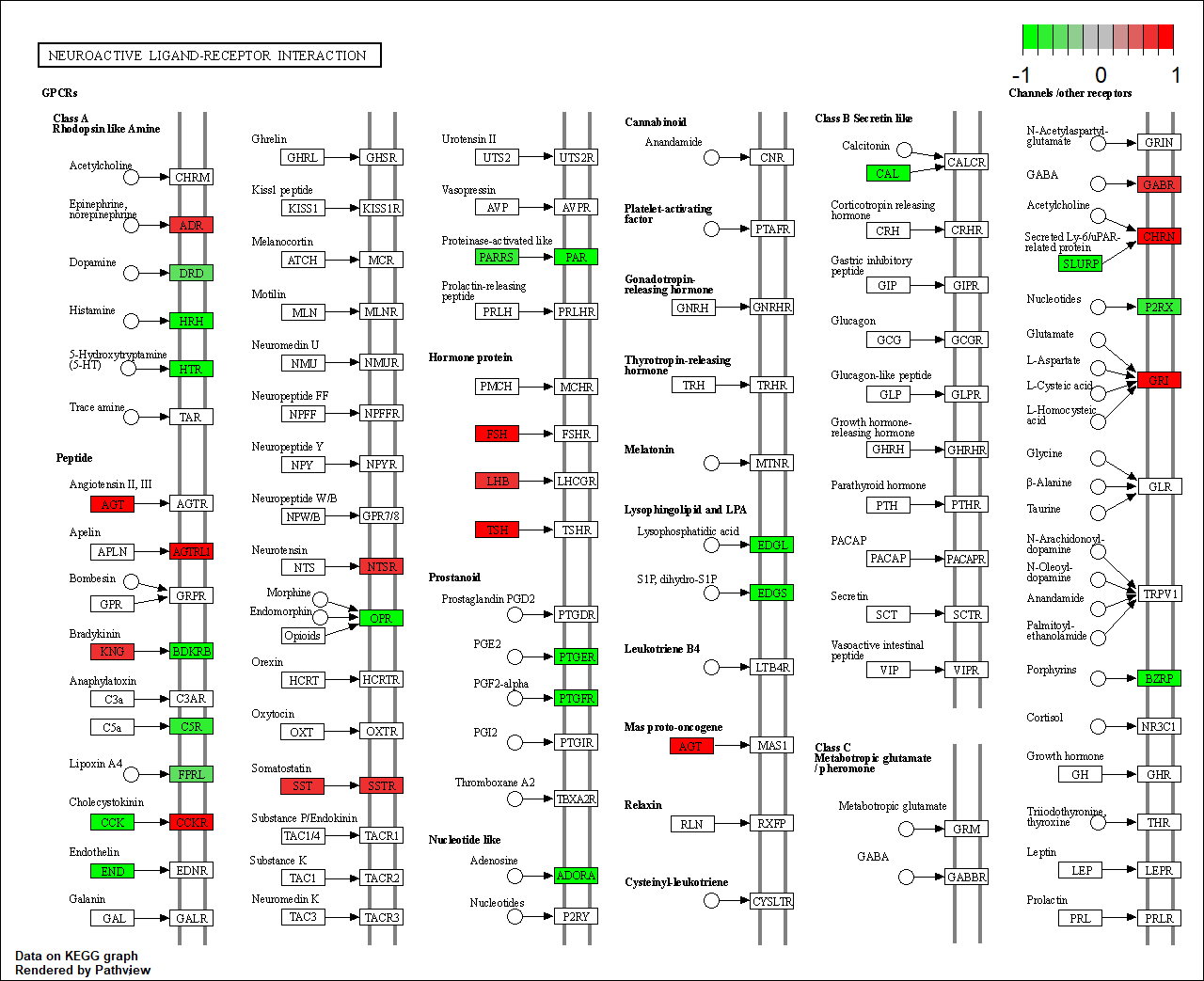
|
|
HTR1D |
hsa04742 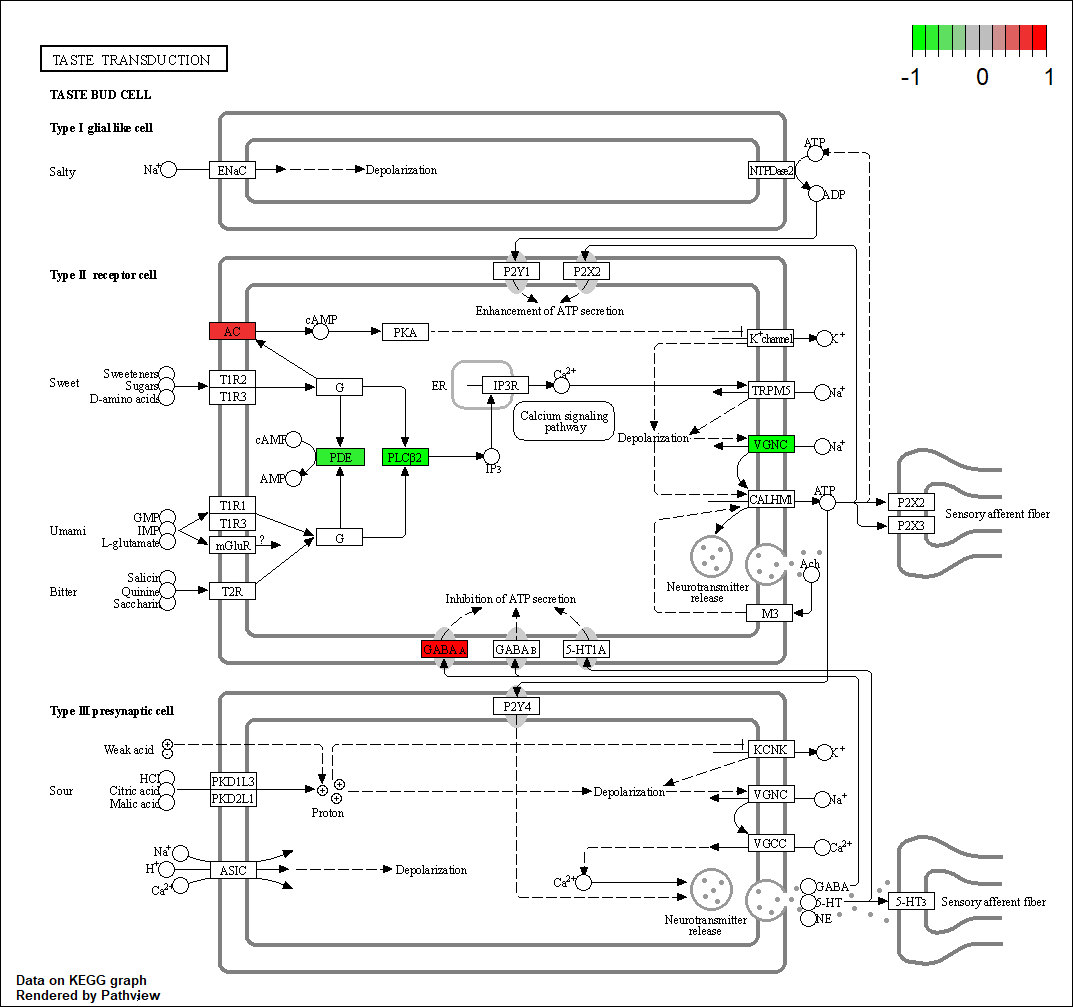
|
|
HTR2A |
hsa04540 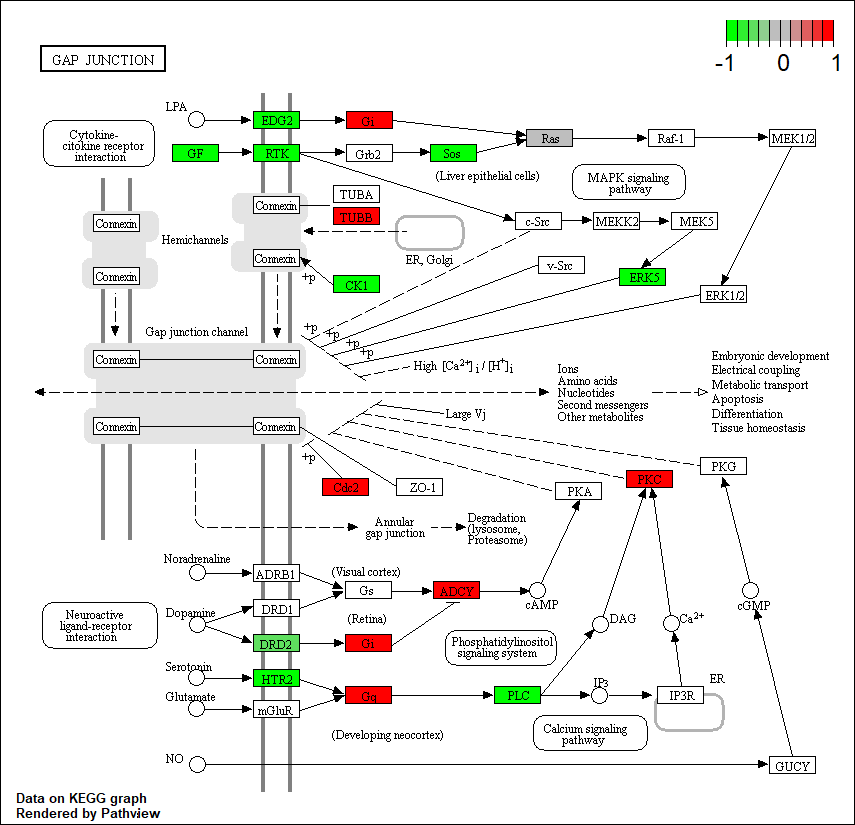
|
|
HTR2B |
hsa04750 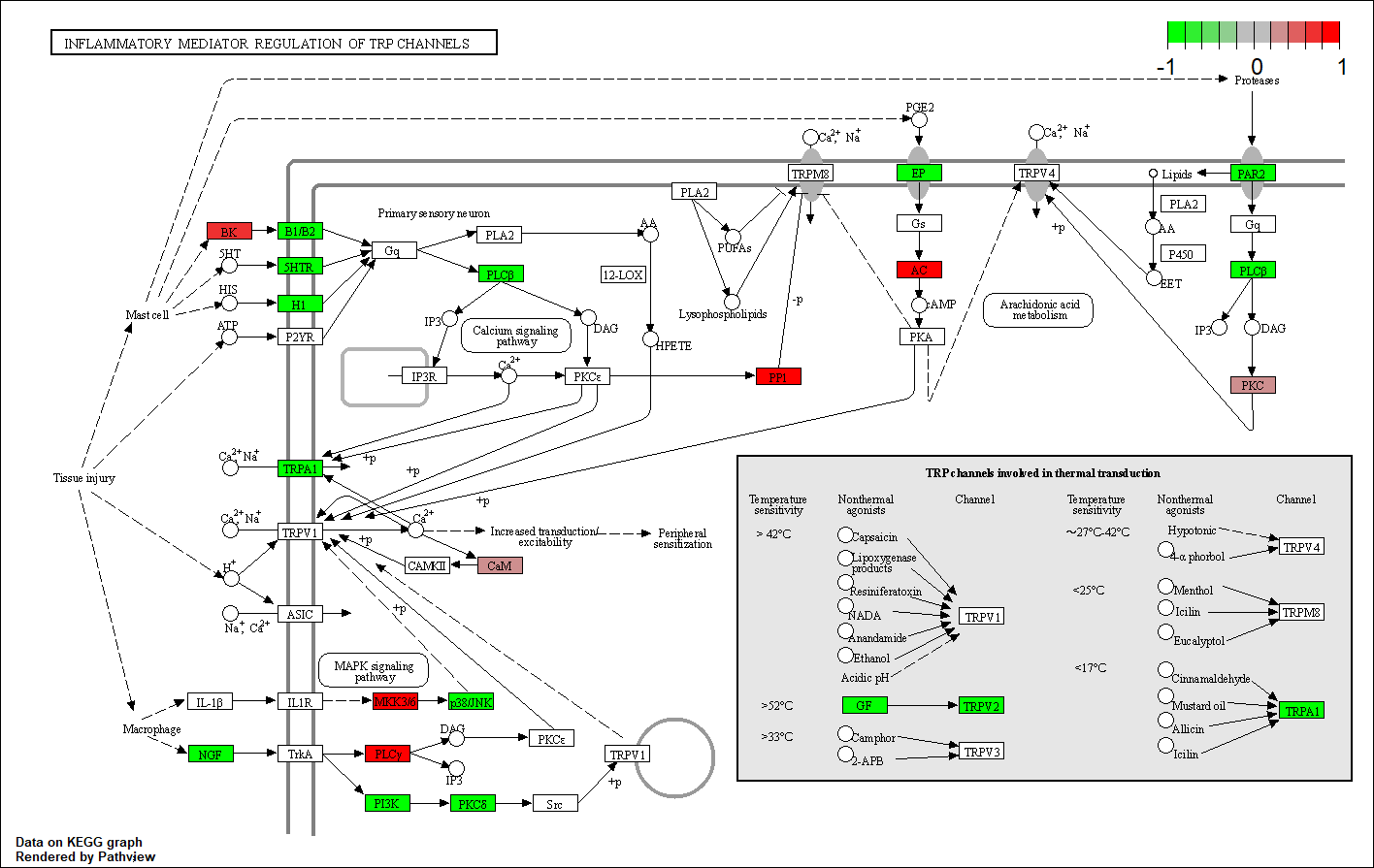
|
|
HTR2C 
|
hsa04726 
|
|
HTR6 |
|
|
HTR7 |
hsa04080 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| FAM169A | 8.4379 | 9.1171 | 8.4463 | 6.94 | 6.7535 | 7.0206 |
| VASH2 | 6.0307 | 5.5989 | 5.5999 | 4.179 | 4.1054 | 4.4684 |
| PROS1 | 9.3287 | 9.3491 | 9.4952 | 5.1828 | 4.7603 | 5.1003 |
| CDK4 | 12.8082 | 12.9604 | 13.353 | 9.1753 | 10.3914 | 10.7191 |
| AURKB | 11.0783 | 10.5393 | 11.3085 | 8.4509 | 8.1853 | 8.4444 |
| AURKA | 7.9629 | 7.9432 | 8.0641 | 5.7957 | 5.2655 | 6.2464 |
| HMGA2 | 11.3612 | 11.4517 | 11.5089 | 8.4324 | 8.6892 | 8.6731 |
| ERBB3 | 10.1175 | 9.3329 | 9.6808 | 7.1418 | 6.3435 | 5.9736 |
| TFDP1 | 9.5679 | 10.3283 | 10.4459 | 8.216 | 7.8494 | 8.1396 |
| PCNA | 14.5819 | 14.5624 | 14.6943 | 13.0044 | 13.1639 | 13.2319 |
| PFKL | 9.1958 | 8.5241 | 9.1478 | 7.3196 | 6.5349 | 6.5765 |
| PLK1 | 13.3609 | 13.2284 | 13.2036 | 11.7141 | 11.0683 | 11.4187 |
| MYLK | 6.001 | 6.0644 | 5.8142 | 4.8516 | 4.3028 | 4.2349 |
| FGFR4 | 12.1166 | 11.4877 | 11.6853 | 6.6346 | 6.6259 | 8.9452 |
| GSTM2 | 5.5849 | 5.3109 | 5.5775 | 4.1248 | 4.1039 | 4.2249 |
| ICAM3 | 7.6834 | 7.4713 | 7.4442 | 5.0458 | 4.2768 | 4.5866 |
| MCM3 | 12.629 | 12.264 | 12.5164 | 10.8958 | 10.9014 | 10.6858 |
| BIRC5 | 14.0991 | 13.7906 | 13.8281 | 12.3395 | 11.9094 | 11.9143 |
| CCNA2 | 11.8502 | 11.635 | 12.0767 | 9.4157 | 9.5595 | 8.8169 |
| RPA1 | 12.0474 | 11.9014 | 12.0247 | 10.6035 | 10.7213 | 10.7478 |
| LIG1 | 10.6459 | 10.3903 | 10.3169 | 6.8944 | 6.5974 | 6.436 |
| BLCAP | 12.2036 | 12.0767 | 12.1286 | 10.4298 | 9.9584 | 9.8391 |
| KIF2C | 11.5535 | 11.0646 | 11.2116 | 9.878 | 9.5702 | 9.5353 |
| B4GAT1 | 12.4692 | 12.2328 | 12.8481 | 9.2592 | 8.9706 | 9.2705 |
| CDK1 | 12.0924 | 11.6808 | 11.5027 | 9.5186 | 9.3677 | 8.8613 |
| GNAI2 | 7.672 | 7.2853 | 7.7848 | 4.9956 | 5.7393 | 5.8153 |
| PIP4K2B | 10.0748 | 9.2994 | 9.6079 | 7.915 | 7.3305 | 7.803 |
| RPA2 | 10.7837 | 10.7144 | 10.6416 | 9.2743 | 9.4545 | 9.3044 |
| BNIP3 | 13.1106 | 12.8928 | 13.1268 | 11.8215 | 11.8309 | 11.6011 |
| MYBL2 | 11.1693 | 10.2445 | 10.8456 | 6.9978 | 7.9385 | 7.6942 |
| CCDC85B | 9.2032 | 9.4712 | 8.9797 | 6.1097 | 6.4048 | 5.7463 |
| DDB2 | 9.6507 | 9.9112 | 9.0347 | 7.0352 | 6.3435 | 6.8049 |
| HADH | 10.8639 | 11.175 | 10.7867 | 6.9052 | 4.9005 | 7.535 |
| STMN1 | 11.6776 | 11.6397 | 11.758 | 7.7854 | 7.5542 | 7.3276 |
| PGM1 | 11.7042 | 11.6475 | 11.7823 | 10.2737 | 10.112 | 10.0544 |
| NET1 | 11.1267 | 11.1003 | 11.0984 | 8.9472 | 9.2808 | 9.3391 |
| PAFAH1B3 | 11.091 | 10.9963 | 11.0359 | 9.2592 | 8.8829 | 8.3493 |
| G3BP1 | 10.01 | 10.1195 | 10.4165 | 7.1976 | 7.7548 | 7.486 |
| CDCA4 | 10.5419 | 10.5697 | 10.2757 | 7.4051 | 6.9095 | 6.6141 |
| NUSAP1 | 11.3012 | 11.5053 | 11.5573 | 9.5199 | 9.3345 | 9.201 |
| PXMP2 | 10.5228 | 10.7553 | 10.6035 | 9.21 | 8.7501 | 9.379 |
| CHEK2 | 10.3956 | 10.1351 | 11.0478 | 8.6892 | 8.282 | 8.5305 |
| ADH5 | 12.6063 | 12.3957 | 12.799 | 11.4604 | 11.1425 | 11.2004 |
| BPHL | 9.4545 | 9.8728 | 9.4614 | 6.3936 | 6.7395 | 6.5351 |
| AMDHD2 | 11.518 | 11.6888 | 11.2862 | 7.4685 | 7.0048 | 6.5242 |
| ACAT2 | 13.0273 | 13.0575 | 12.6895 | 9.7104 | 10.5323 | 10.891 |
| PIN1 | 13.4289 | 13.6203 | 13.5245 | 11.8381 | 11.9252 | 12.2254 |
| RPIA | 13.6124 | 13.5637 | 13.7667 | 12.13 | 12.1533 | 12.336 |
| RFC2 | 9.5831 | 9.1262 | 9.5867 | 7.5818 | 7.7366 | 7.6309 |
| ADI1 | 10.6008 | 10.1569 | 10.5471 | 8.595 | 9.0813 | 8.7241 |
| TSPAN4 | 11.6947 | 11.5766 | 11.6165 | 10.4225 | 10.4225 | 10.1524 |
| GPER1 | 8.8345 | 8.6832 | 8.9105 | 6.9438 | 6.8408 | 6.311 |
| POLE2 | 9.1992 | 9.354 | 9.1659 | 7.7098 | 7.1747 | 6.9297 |
| SLC37A4 | 7.4723 | 7.5992 | 7.8464 | 5.3106 | 4.9848 | 4.9522 |
| DLGAP5 | 11.1854 | 11.2204 | 11.4665 | 9.0564 | 8.0154 | 8.4112 |
| FAM162A | 11.862 | 12.217 | 11.634 | 9.49 | 8.6484 | 9.1588 |
| CEP55 | 8.7246 | 8.4678 | 8.6213 | 6.8022 | 6.7255 | 6.7693 |
| GLUL | 8.2339 | 8.3728 | 7.5773 | 2.436 | 2.4785 | 2.4758 |
| CTSA | 14.1184 | 14.2757 | 13.8723 | 11.9074 | 12.1241 | 11.6858 |
| SUMO3 | 11.4784 | 11.2471 | 10.7083 | 8.9665 | 8.7106 | 8.6236 |
| NREP | 5.9582 | 5.0619 | 5.4411 | 2.8622 | 3.3497 | 3.3437 |
| IFITM2 | 11.404 | 10.4542 | 11.079 | 8.0949 | 7.944 | 7.3063 |
| GLB1 | 10.8754 | 10.0741 | 10.1972 | 7.8931 | 8.2681 | 8.0877 |
| LANCL1 | 9.5877 | 9.6385 | 9.7125 | 7.594 | 7.6837 | 7.8063 |
| FIBP | 9.4895 | 9.3126 | 9.1486 | 8.1551 | 8.0524 | 7.93 |
| UNG | 11.5036 | 11.1843 | 11.4009 | 7.8877 | 9.122 | 8.8948 |
| TK1 | 10.4321 | 10.1184 | 10.0709 | 8.0609 | 8.5518 | 8.5438 |
| DHFR | 10.4664 | 10.2472 | 9.8291 | 8.3314 | 8.4714 | 8.1829 |
| TOB1 | 10.6154 | 10.2518 | 10.6246 | 9.3173 | 8.7778 | 8.9518 |
| RNASEH2A | 10.8449 | 10.8165 | 11.0484 | 9.2341 | 8.5076 | 8.0674 |
| CYBA | 10.2508 | 10.3245 | 10.814 | 7.7657 | 7.9444 | 8.2122 |
| SPAG5 | 10.7422 | 10.4247 | 10.1392 | 7.5051 | 8.2007 | 8.6508 |
| TMPO | 11.8701 | 12.2098 | 12.0038 | 10.2086 | 10.6101 | 10.2938 |
| SKP2 | 12.4847 | 12.2651 | 12.8774 | 9.1882 | 9.309 | 8.6171 |
| NQO2 | 12.6012 | 12.5835 | 12.6715 | 10.8018 | 10.107 | 10.5723 |
| VRK1 | 9.69 | 10.0131 | 9.9262 | 8.0925 | 8.4105 | 7.4701 |
| TRIP13 | 10.8958 | 10.5905 | 10.8549 | 8.2813 | 8.632 | 7.8479 |
| MYL6B | 13.2896 | 12.4928 | 12.7722 | 9.7541 | 10.3706 | 10.1138 |
| GTSE1 | 10.1077 | 10.0505 | 9.4295 | 7.5789 | 7.6933 | 8.0003 |
| POLA2 | 8.1332 | 7.7036 | 7.7435 | 6.3131 | 6.366 | 6.5471 |
| KIF11 | 9.8715 | 9.6119 | 9.6627 | 7.0192 | 7.5923 | 8.0372 |
| KIF23 | 8.9959 | 9.6438 | 9.3284 | 7.3311 | 7.416 | 7.804 |
| SLC29A2 | 7.8075 | 8.1667 | 7.9098 | 6.7351 | 6.1519 | 6.2763 |
| NUDT1 | 7.9406 | 7.2825 | 7.2738 | 5.6428 | 4.7771 | 5.1096 |
| MGMT | 8.8103 | 8.7413 | 8.1023 | 7.0097 | 6.6135 | 6.817 |
| SH3BGR | 5.351 | 5.3587 | 5.4567 | 4.2884 | 3.8609 | 4.0955 |
| RAD51 | 9.1988 | 9.1843 | 8.9685 | 6.6172 | 7.5287 | 7.3383 |
| PRIM1 | 8.6949 | 8.6695 | 8.5629 | 6.3125 | 7.1858 | 6.9828 |
| CDC25C | 8.0584 | 8.5181 | 8.1908 | 6.7004 | 6.0956 | 6.1634 |
| PEBP1 | 14.6275 | 14.0091 | 14.4593 | 12.4229 | 10.9822 | 11.5589 |
| GAMT | 10.1787 | 9.5641 | 10.0587 | 7.2979 | 7.4866 | 6.7403 |
| PDZK1 | 8.709 | 9.5169 | 8.4094 | 5.7755 | 4.3413 | 4.9732 |
| SAC3D1 | 11.2538 | 11.1243 | 10.9388 | 9.124 | 9.4117 | 8.7279 |
| SPINK2 | 7.5358 | 7.2241 | 7.1539 | 5.0674 | 5.7129 | 5.1188 |
| CENPF | 9.2698 | 9.3747 | 8.9982 | 7.2837 | 6.8943 | 6.9705 |
| CD164 | 13.2976 | 13.3573 | 13.6583 | 11.5844 | 11.9014 | 11.6786 |
| C1S | 8.413 | 8.0745 | 8.7274 | 6.3209 | 5.5727 | 5.4185 |
| XPO1 | 12.3803 | 12.4668 | 12.8043 | 10.2854 | 10.0889 | 10.6711 |
| CCNG1 | 10.8796 | 10.2209 | 11.0008 | 7.4852 | 8.4553 | 8.2078 |
| DUT | 11.5124 | 11.5901 | 11.661 | 9.4319 | 10.1324 | 9.1746 |
| TUBB | 14.992 | 15 | 14.9161 | 12.8164 | 12.3702 | 12.5783 |
| QDPR | 10.6681 | 11.3953 | 10.7478 | 8.1175 | 5.5802 | 6.7448 |
| POLR2H | 10.8333 | 10.7406 | 10.7917 | 9.3057 | 9.1482 | 8.7386 |
| AMACR | 8.2451 | 9.6518 | 8.3901 | 6.0492 | 5.0582 | 5.0598 |
| LRP5 | 10.1568 | 10.4131 | 10.3772 | 8.8063 | 8.7768 | 9.1635 |
| NSL1 | 10.1222 | 10.4291 | 10.3357 | 9.0494 | 8.9859 | 8.6062 |
| PCYT2 | 11.3532 | 11.6288 | 11.4833 | 9.9118 | 9.4754 | 9.4193 |
| BUB1 | 11.4331 | 11.2493 | 11.4241 | 9.479 | 9.6498 | 8.9904 |
| TTR | 10.7176 | 11.5819 | 11.2478 | 8.9905 | 9.037 | 8.8896 |
| SPC25 | 9.4867 | 10.0099 | 9.7114 | 6.8221 | 7.0503 | 7.6639 |
| ABHD14A | 8.8253 | 8.9744 | 8.8984 | 6.9536 | 6.9997 | 7.1938 |
| TPX2 | 13.6853 | 12.9548 | 13.9396 | 11.4122 | 10.8922 | 10.9818 |
| BIN1 | 7.6346 | 7.4324 | 7.4185 | 5.8657 | 5.6004 | 5.3751 |
| SIVA1 | 11.9252 | 11.2769 | 11.5073 | 9.8524 | 9.982 | 9.99 |
| KHK | 9.1709 | 8.851 | 9.0307 | 7.5833 | 7.5166 | 7.1888 |
| HSPA2 | 8.7599 | 9.1302 | 9.3364 | 5.7645 | 6.4444 | 5.68 |
| ARL6IP1 | 12.7491 | 12.6222 | 13.0677 | 10.7531 | 11.3364 | 11.2645 |
| MKI67 | 8.9045 | 8.3405 | 8.6669 | 7.1161 | 6.8476 | 6.8415 |
| IFITM3 | 13.0015 | 13.6217 | 13.1064 | 9.0512 | 10.5813 | 8.9536 |
| NME4 | 12.7569 | 12.6936 | 13.2481 | 8.8419 | 9.0598 | 8.8864 |
| GTF2H5 | 11.5384 | 11.7594 | 11.4923 | 9.3448 | 9.2926 | 9.5333 |
| ZBTB1 | 7.6214 | 8.1908 | 8.282 | 6.5741 | 6.5483 | 6.5064 |
| CHN2 | 8.9066 | 9.3272 | 9.135 | 7.1786 | 7.432 | 7.4813 |
| KAZN | 13.5963 | 13.4774 | 13.2565 | 10.4002 | 11.1773 | 10.2699 |
| OIP5 | 10.0027 | 9.8755 | 9.7202 | 7.4558 | 7.668 | 7.8838 |
| LGR5 | 11.1739 | 12.4906 | 12.6854 | 8.7314 | 9.2305 | 9.602 |
| SRSF7 | 11.7789 | 11.2654 | 11.9074 | 9.9143 | 9.9348 | 9.5475 |
| POLE | 9.4446 | 8.8655 | 8.5181 | 6.558 | 6.6705 | 7.0728 |
| RBBP4 | 12.0683 | 12.2302 | 12.2927 | 10.8043 | 10.4713 | 10.7077 |
| MTCH2 | 13.888 | 13.137 | 13.0545 | 11.0156 | 11.1582 | 11.5337 |
| MRPL16 | 12.8401 | 13.1768 | 12.9182 | 10.8368 | 11.3922 | 11.4812 |
| FKBP3 | 14.0733 | 13.2319 | 13.8646 | 11.7918 | 11.8823 | 11.9187 |
| ECI2 | 14.6135 | 13.9475 | 14.3105 | 11.9209 | 10.4651 | 10.8428 |
| ASF1B | 10.0544 | 10.0679 | 10.3827 | 7.5684 | 7.9973 | 7.3265 |
| SPATA20 | 11.3942 | 11.2004 | 10.7057 | 9.4322 | 9.4326 | 9.5934 |
| SLC25A10 | 9.3843 | 8.915 | 9.4712 | 7.7289 | 7.6087 | 7.7792 |
| CARHSP1 | 11.753 | 12.7446 | 12.0145 | 10.0615 | 9.2019 | 8.8505 |
| SNX10 | 9.2608 | 9.7502 | 9.8469 | 6.8862 | 6.7312 | 6.0068 |
| KANK2 | 8.1908 | 8.5567 | 8.1203 | 6.5315 | 6.5293 | 6.0651 |
| TMEM14A | 9.8342 | 9.5077 | 10.5964 | 6.1308 | 6.2754 | 6.8931 |
| SNRNP25 | 13.2381 | 12.9277 | 12.9988 | 9.9387 | 9.8949 | 10.2714 |
| SLC2A4RG | 10.284 | 10.4428 | 11.0999 | 8.3473 | 8.2992 | 8.3657 |
| HILPDA | 8.2527 | 8.0968 | 8.2421 | 6.4789 | 6.7986 | 6.3793 |
| MRS2 | 11.4647 | 11.5249 | 11.8336 | 10.019 | 10.017 | 10.013 |
| FZD4 | 9.3704 | 8.9271 | 8.9521 | 7.6935 | 7.7768 | 7.4836 |
| RAB29 | 8.7481 | 8.9797 | 9.0102 | 7.2115 | 7.4256 | 7.5368 |
| PAAF1 | 8.5896 | 8.5745 | 8.0829 | 6.9845 | 6.7498 | 6.6609 |
| MIS18A | 9.426 | 8.9757 | 8.9434 | 7.5351 | 7.7214 | 7.3493 |
| GALNT11 | 9.1977 | 8.6707 | 8.8616 | 7.3867 | 7.208 | 6.9894 |
| SIRT5 | 8.1216 | 8.5542 | 8.0975 | 6.4696 | 5.8064 | 6.0639 |
| MACROD1 | 10.4447 | 10.2377 | 10.4641 | 8.4655 | 7.9754 | 8.4064 |
| ELP5 | 11.71 | 12.13 | 11.9578 | 10.3056 | 10.2456 | 10.1368 |
| APOA2 | 13.6536 | 13.7274 | 13.5063 | 12.1581 | 12.3022 | 11.6531 |
| DECR2 | 8.7796 | 8.8573 | 8.6159 | 7.4847 | 6.9669 | 6.9551 |
| E2F8 | 6.754 | 7.0127 | 7.03 | 4.2656 | 5.1844 | 5.0909 |
| AUNIP | 7.6744 | 8.2741 | 8.3636 | 6.4296 | 6.5539 | 6.1956 |
| RMND1 | 9.972 | 10.3148 | 10.2476 | 8.5507 | 8.2402 | 8.556 |
| NELFCD | 12.0749 | 12.7741 | 12.5403 | 10.4685 | 10.7601 | 10.6224 |
| OSGEPL1 | 6.9187 | 6.5264 | 6.9572 | 5.337 | 5.3283 | 5.0876 |
| DNMT3B | 11.0698 | 10.5151 | 10.2101 | 8.5273 | 8.1624 | 8.3242 |
| C1QTNF3 | 6.1157 | 6.5714 | 6.1375 | 3.7156 | 3.5055 | 3.8589 |
| CDCA3 | 11.6475 | 12.4403 | 11.9435 | 9.4797 | 9.4267 | 9.4885 |
| GINS2 | 11.1992 | 10.4443 | 10.6214 | 8.0706 | 8.8103 | 8.3134 |
| CHPT1 | 13.2865 | 13.0761 | 12.9107 | 10.8333 | 10.3013 | 10.9499 |
| SUB1 | 12.159 | 12.3968 | 12.0571 | 10.8202 | 10.9379 | 10.9545 |
| GLRX5 | 11.2505 | 11.371 | 10.7061 | 8.6485 | 7.0541 | 8.5131 |
| WRAP53 | 9.3659 | 9.1005 | 9.1154 | 7.9722 | 7.7244 | 8.006 |
| HCFC1R1 | 9.7745 | 9.905 | 9.8847 | 8.3707 | 8.1695 | 8.0448 |
| PAH | 8.9781 | 9.0819 | 8.761 | 7.2302 | 6.961 | 7.6162 |
| MAT1A | 7.7415 | 7.4763 | 7.6342 | 6.5123 | 6.4125 | 6.4107 |
| MYH3 | 7.7585 | 7.7717 | 7.8895 | 6.2055 | 6.3488 | 6.3369 |
| LEFTY1 | 7.5301 | 7.2942 | 7.2807 | 5.213 | 5.6645 | 5.9721 |
| F7 | 7.8468 | 7.5454 | 7.6078 | 6.2512 | 6.323 | 6.1342 |
SMARCA4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: SMARCA4
[1] SMARCA4-Deficient Thoracic Sarcoma: A Case Report and Review of Literature.
[2] [SMARCA4-deficient thoracic tumors: A new entity].
[3] SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non-small cell lung cancer.
[5] SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: a multi-cohort analysis.
[8] Recurrent Loss of SMARCA4 in Sinonasal Teratocarcinosarcoma.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa04714
