KLF10 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN3189
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: AR-A014418
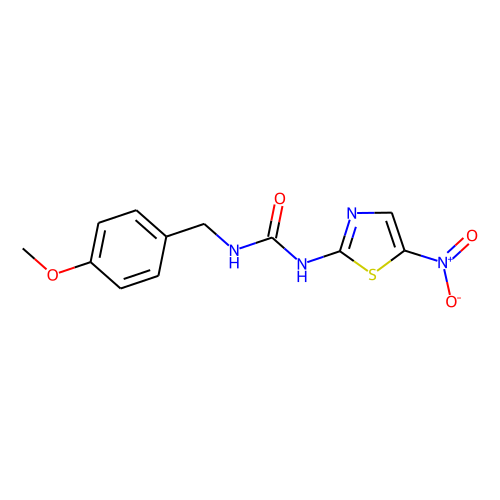
| ID | BRD-K67860401 |
| name | AR-A014418 |
| type | Small molecule |
| molecular formula | C12H12N4O4S |
| molecular weight | 308.32 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | COc1ccc(CNC(=O)Nc2ncc(s2)[N+]([O-])=O)cc1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: HEPG2
Cell lines used for experiments
TRANSCRIPTION FACTOR: KLF10
Transcription regulatory factors corresponding to this network
ARTICLE: AR-A014418
[1] GSK3β suppression inhibits MCL1 protein synthesis in human acute myeloid leukemia cells.
[2] N-(4-Meth-oxy-phen-yl)-N'-(5-nitro-1,3-thia-zol-2-yl)urea.
[4] GSK3α/β: A Novel Therapeutic Target for Neuroendocrine Tumors.
[9] Inhibition of GSK-3β on Behavioral Changes and Oxidative Stress in an Animal Model of Mania.
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
GSK3B 
|
hsa04910 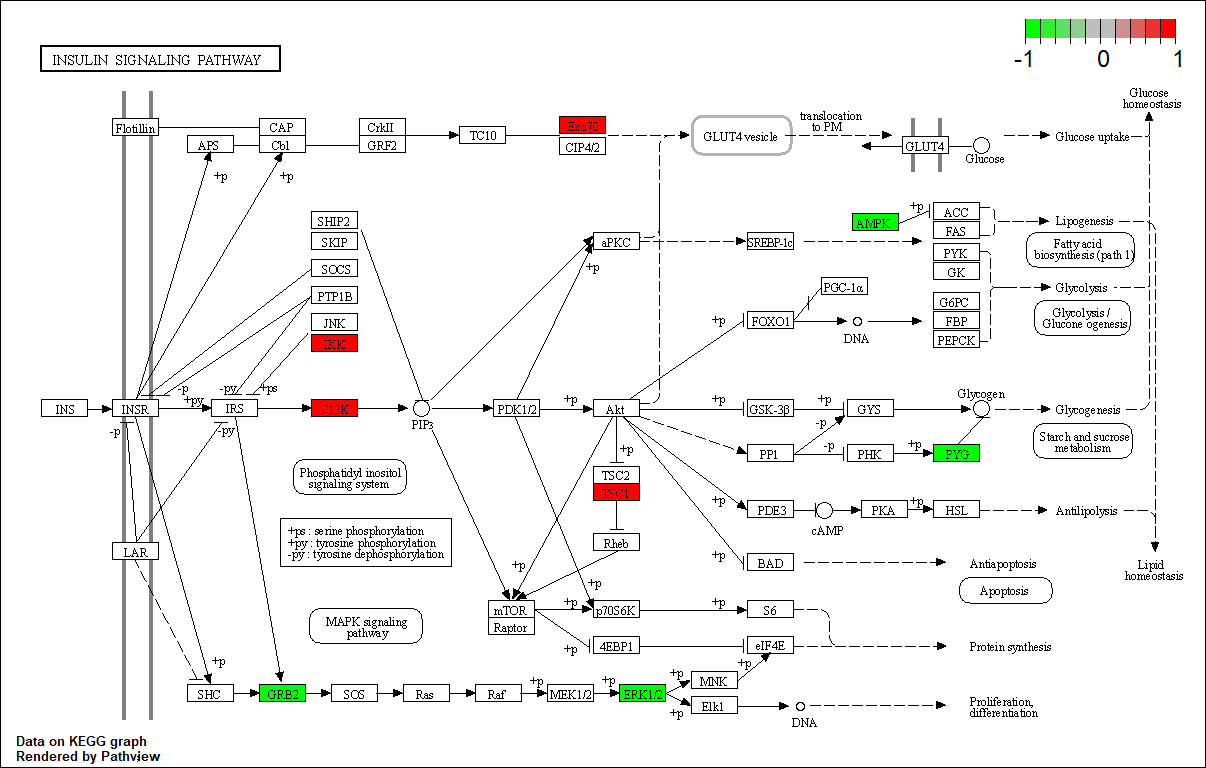
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| ENTPD1 | 5.6672 | 6.9122 | 6.1539 | 4.2348 | 4.8687 | 4.3705 |
| L3MBTL1 | 7.3933 | 6.9927 | 7.1563 | 5.8957 | 5.629 | 5.6706 |
| CEP68 | 5.8121 | 5.403 | 5.5429 | 4.7306 | 3.9587 | 4.5599 |
| PCGF3 | 8.1267 | 8.4988 | 8.4002 | 6.7823 | 7.0694 | 6.9853 |
| CLEC16A | 7.1934 | 7.4244 | 7.656 | 6.6448 | 6.411 | 6.5712 |
| CBX7 | 5.4907 | 5.4716 | 4.9816 | 4.0791 | 4.2699 | 4.1495 |
| SALL2 | 9.4309 | 8.8108 | 8.9246 | 7.5234 | 7.5779 | 6.7133 |
| ZNF500 | 7.0651 | 7.1367 | 6.6973 | 6.0352 | 5.9447 | 6.2073 |
| METTL3 | 7.452 | 7.1466 | 7.304 | 4.9838 | 5.8837 | 5.7984 |
| GTF2H2B | 7.9209 | 6.8735 | 7.3188 | 5.6179 | 5.1656 | 5.1765 |
| ZSCAN32 | 6.8701 | 6.7343 | 6.6769 | 5.9371 | 6.023 | 5.465 |
| DCUN1D2 | 7.3422 | 7.4008 | 7.2572 | 6.3066 | 6.4899 | 6.6594 |
| ZNF232 | 10.2535 | 10.7334 | 10.4777 | 8.6873 | 9.013 | 8.3921 |
| TRMT13 | 7.1627 | 7.2921 | 7.1339 | 5.3515 | 5.4004 | 5.2522 |
| PTCD2 | 4.8213 | 4.6909 | 4.6754 | 4.0108 | 3.8541 | 4.0174 |
| CDADC1 | 6.0206 | 5.8876 | 6.1329 | 5.0676 | 5.1142 | 5.1744 |
| CSAD | 6.2691 | 6.3234 | 6.301 | 5.5703 | 4.8604 | 5.2537 |
| TARDBP | 9.3435 | 8.3995 | 9.3435 | 7.1789 | 6.5626 | 7.6146 |
| TBC1D12 | 8.079 | 7.7444 | 7.8911 | 6.8611 | 7.0863 | 7.1435 |
| NSUN6 | 5.998 | 6.1965 | 6.0287 | 5.0248 | 5.4243 | 5.1126 |
| DFFB | 6.6053 | 6.2494 | 6.6076 | 5.3252 | 5.4103 | 5.816 |
| PIK3R4 | 8.1497 | 8.039 | 8.0433 | 7.0442 | 6.972 | 6.4404 |
| AURKA | 8.2633 | 7.9615 | 7.8103 | 7.0945 | 7.0446 | 7.1551 |
| PIK3R3 | 5.3688 | 5.1069 | 4.9763 | 4.211 | 4.2163 | 4.335 |
| PLK1 | 13.5794 | 13.5088 | 13.5037 | 12.1459 | 12.4764 | 11.7076 |
| APPBP2 | 8.2633 | 8.3146 | 8.5119 | 7.2922 | 6.6546 | 6.5055 |
| EFCAB14 | 8.3643 | 8.5724 | 8.2808 | 7.176 | 7.0358 | 7.4601 |
| TXLNA | 14.0164 | 13.8117 | 13.8972 | 12.4327 | 12.5635 | 13.1613 |
| TRAK2 | 7.4489 | 7.0685 | 7.3426 | 5.3252 | 5.1086 | 5.5488 |
| BUB1B | 11.4196 | 11.6506 | 11.4249 | 10.3314 | 10.2759 | 9.9426 |
| ENOSF1 | 6.7545 | 6.5081 | 6.8597 | 5.9433 | 5.4265 | 5.5792 |
| PAFAH1B1 | 9.3655 | 9.4434 | 9.5555 | 8.3026 | 7.701 | 7.6881 |
| PSRC1 | 10.3531 | 10.1141 | 10.2785 | 7.6701 | 8.8973 | 7.5543 |
| NUP62 | 10.4608 | 10.3723 | 9.9371 | 9.0324 | 8.831 | 7.8802 |
| KIF2C | 12.0565 | 11.6616 | 11.6349 | 10.3867 | 10.3512 | 10.2313 |
| PIP4K2B | 9.5397 | 10.1717 | 9.6299 | 8.3691 | 8.1699 | 7.0181 |
| STMN1 | 12.4135 | 11.6079 | 11.9419 | 10.8542 | 10.8752 | 10.6805 |
| TSEN2 | 8.8132 | 8.1837 | 7.8625 | 6.8689 | 6.4303 | 6.3229 |
| IKBKE | 6.0788 | 5.9279 | 6.1668 | 4.9549 | 5.3321 | 5.2599 |
| NUSAP1 | 12.2065 | 11.9569 | 12.0959 | 10.9416 | 10.7421 | 10.4821 |
| GPATCH8 | 7.9299 | 8.4115 | 8.4426 | 6.7846 | 6.6706 | 6.7283 |
| EIF5 | 13.1079 | 12.9232 | 12.9708 | 11.0175 | 10.7831 | 10.9772 |
| LSR | 11.6357 | 11.7672 | 11.7066 | 9.8405 | 10.0147 | 9.8883 |
| ZNF589 | 6.9667 | 6.9777 | 7.2123 | 5.0855 | 5.463 | 5.8916 |
| ZDHHC6 | 9.4408 | 9.3094 | 9.5371 | 8.173 | 8.2612 | 7.7071 |
| THAP11 | 9.399 | 9.0412 | 9.2426 | 8.4012 | 8.1002 | 8.3743 |
| PARP2 | 11.5944 | 11.575 | 11.6142 | 8.0106 | 8.5137 | 7.7339 |
| KIAA0753 | 7.9452 | 7.5905 | 7.8914 | 6.5372 | 6.5708 | 6.22 |
| ABHD6 | 7.5774 | 7.2399 | 7.3505 | 6.4831 | 6.1424 | 6.1897 |
| RPP38 | 8.0019 | 8.0524 | 7.9661 | 7.2483 | 7.0766 | 7.388 |
| CCNF | 8.8427 | 8.7456 | 8.2679 | 6.3129 | 6.4098 | 6.6895 |
| ZC3H4 | 8.8132 | 8.9314 | 8.5306 | 7.5076 | 7.4322 | 7.9063 |
| FZD5 | 13.0735 | 13.101 | 12.5728 | 11.3088 | 11.3249 | 10.9877 |
| DDX1 | 13.0746 | 13.5037 | 13.0691 | 12.4699 | 12.0559 | 12.0204 |
| CUL4A | 11.1016 | 11.4782 | 10.6883 | 9.8553 | 10.0658 | 9.9451 |
| TIA1 | 10.3036 | 11.1386 | 10.2322 | 8.6908 | 9.1703 | 8.7277 |
| ZFR | 11.3257 | 10.9377 | 11.3179 | 10.2796 | 9.9868 | 10.3981 |
| PPP2R5C | 8.5476 | 8.3071 | 8.1209 | 7.176 | 6.5907 | 7.055 |
| RRM2 | 14.0484 | 13.7602 | 13.2578 | 11.9517 | 11.9414 | 12.4376 |
| ZMYM4 | 9.6401 | 9.0392 | 9.7343 | 8.5156 | 8.2444 | 7.9187 |
| SEL1L | 10.661 | 10.5531 | 10.2358 | 9.6975 | 9.2394 | 8.9051 |
| MTX2 | 12.3182 | 12.1411 | 12.4227 | 11.3367 | 11.2791 | 11.4021 |
| SKP2 | 12.025 | 12.374 | 12.401 | 10.8157 | 10.3971 | 11.4324 |
| CSTF3 | 9.3569 | 9.4149 | 9.3943 | 8.4711 | 8.323 | 7.8601 |
| MEIS1 | 8.2434 | 8.4408 | 7.8346 | 5.9905 | 5.9097 | 5.2674 |
| RFC3 | 12.9635 | 12.3663 | 13.0859 | 11.5689 | 11.2391 | 11.6349 |
| COL7A1 | 6.6154 | 6.8157 | 6.667 | 5.1993 | 5.0297 | 5.9846 |
| EFNA4 | 9.2193 | 9.1197 | 8.981 | 8.2757 | 8.469 | 8.2928 |
| BARD1 | 8.0866 | 8.0573 | 8.0261 | 7.3358 | 6.835 | 6.7225 |
| MLLT10 | 7.7122 | 8.2685 | 8.389 | 7.0419 | 7.1901 | 6.6788 |
| GEMIN4 | 10.0845 | 10.026 | 10.3314 | 8.9449 | 9.265 | 9.2916 |
| OGG1 | 9.1677 | 9.2406 | 9.1031 | 8.5213 | 8.1591 | 8.2788 |
| GINS1 | 12.7717 | 12.6827 | 12.8668 | 11.2765 | 10.9861 | 10.9914 |
| SLC16A5 | 5.7177 | 5.2457 | 5.7864 | 4.5925 | 4.6218 | 4.4587 |
| LETMD1 | 9.9177 | 10.0359 | 9.7812 | 8.902 | 8.7818 | 8.7698 |
| EXOSC10 | 12.0825 | 12.3311 | 12.3876 | 11.577 | 11.4978 | 11.4401 |
| XPO1 | 12.0511 | 12.2385 | 11.9158 | 11.3483 | 11.2832 | 11.1701 |
| NCOA6 | 9.3312 | 9.5641 | 9.5463 | 8.6044 | 8.6121 | 8.3867 |
| CLN3 | 8.7692 | 8.6858 | 8.7101 | 7.8421 | 7.6396 | 7.8456 |
| BCL2L2 | 11.3692 | 11.1639 | 11.5066 | 10.3747 | 10.5578 | 9.9058 |
| NSL1 | 10.2031 | 9.6645 | 9.9149 | 9.1381 | 8.9044 | 8.9306 |
| EXTL2 | 9.4164 | 10.069 | 9.824 | 8.1221 | 8.6978 | 7.6089 |
| DCLRE1A | 6.6943 | 6.2584 | 6.481 | 5.4101 | 5.2797 | 5.7519 |
| CCNO | 4.9528 | 5.0274 | 5.3999 | 4.1158 | 3.9183 | 3.7721 |
| APEX1 | 15 | 15 | 15 | 13.4108 | 13.763 | 13.359 |
| CROCCP2 | 10.2338 | 9.5821 | 10.1911 | 8.8487 | 8.7771 | 8.6502 |
| CBX5 | 9.3692 | 9.4813 | 9.3977 | 8.0782 | 7.7162 | 7.9053 |
| MRPS31 | 11.6605 | 11.0702 | 11.4933 | 10.4524 | 10.3938 | 10.2036 |
| TBC1D2B | 7.103 | 7.1887 | 7.1238 | 6.1304 | 6.3492 | 6.3123 |
| NFATC2IP | 9.6177 | 9.1128 | 9.1474 | 8.499 | 8.2746 | 8.0696 |
| DICER1 | 8.8983 | 9.6394 | 9.5118 | 7.9083 | 8.0317 | 8.1849 |
| ANKRD28 | 4.3536 | 4.7556 | 4.2398 | 3.503 | 3.426 | 3.2218 |
| RALGAPA1 | 10.2603 | 10.0289 | 10.1378 | 8.7637 | 8.4028 | 8.877 |
| EDRF1 | 8.151 | 7.6823 | 8.5469 | 5.22 | 5.6194 | 5.7661 |
| CBR4 | 9.1383 | 8.8897 | 8.167 | 7.2049 | 6.8515 | 7.5341 |
| PMS1 | 8.4124 | 8.7485 | 8.9951 | 7.319 | 6.4 | 6.708 |
| UPF3A | 10.6082 | 9.9555 | 9.9099 | 8.5945 | 9.1131 | 8.3838 |
| ZNF337 | 7.462 | 7.0297 | 7.2183 | 6.1265 | 6.1864 | 6.2337 |
| SLC7A8 | 6.2231 | 6.483 | 6.3975 | 5.3143 | 5.2786 | 4.7434 |
| SLC35E2B | 6.8235 | 6.1532 | 6.6187 | 5.229 | 4.6638 | 5.2646 |
| EPS15 | 7.2174 | 7.7339 | 7.6817 | 5.9171 | 5.4911 | 6.1779 |
| FKBP3 | 13.6066 | 13.8257 | 13.7544 | 12.018 | 12.3212 | 12.2198 |
| PRPF38B | 7.9136 | 8.0752 | 7.9003 | 7.0158 | 6.8479 | 7.351 |
| ARGLU1 | 10.164 | 9.7397 | 9.6979 | 8.6408 | 8.5502 | 8.8206 |
| METTL17 | 14.0277 | 13.6994 | 13.687 | 12.4891 | 12.0711 | 12.254 |
| S100PBP | 7.4792 | 7.6247 | 7.2165 | 6.2353 | 6.2406 | 5.6384 |
| RBM26 | 12.0541 | 11.4466 | 11.3124 | 10.4621 | 10.4666 | 10.263 |
| BBS1 | 6.8088 | 6.4707 | 7.2982 | 5.8592 | 5.5728 | 5.5587 |
| SMG8 | 9.0108 | 9.1255 | 9.1933 | 8.4547 | 7.9066 | 8.2111 |
| PCTP | 9.1808 | 9.5904 | 9.8253 | 8.1305 | 8.4659 | 8.3117 |
| NCKIPSD | 7.8646 | 7.9497 | 7.652 | 6.7103 | 6.9265 | 7.1622 |
| RNF111 | 7.0213 | 6.4975 | 7.0082 | 5.9752 | 5.9168 | 5.7025 |
| NUP107 | 12.0033 | 12.3528 | 11.7162 | 10.9812 | 11.0529 | 10.9396 |
| FASTKD1 | 8.8241 | 9.0437 | 9.2022 | 8.2928 | 7.8462 | 7.782 |
| C2orf42 | 6.8527 | 6.6202 | 6.8969 | 5.8033 | 5.5244 | 5.4123 |
| MFF | 10.2383 | 10.1329 | 10.122 | 8.9833 | 8.3962 | 8.607 |
| DNAAF2 | 9.1176 | 9.1313 | 9.1914 | 8.345 | 8.2566 | 8.5438 |
| RIC8B | 5.4903 | 6.0589 | 5.655 | 4.6765 | 4.7771 | 4.7738 |
| BORA | 9.0232 | 9.0727 | 9.0371 | 8.0531 | 8.1559 | 7.885 |
| ALG6 | 9.9493 | 9.83 | 10.5553 | 8.7269 | 8.493 | 8.4517 |
| NARF | 11.0964 | 11.0255 | 11.017 | 10.3619 | 10.1069 | 10.2944 |
| ZFYVE21 | 7.4762 | 7.743 | 7.6377 | 6.5774 | 6.8611 | 6.676 |
| HELLS | 7.9748 | 7.7105 | 7.9323 | 6.5226 | 6.687 | 6.3583 |
| ENGASE | 8.2084 | 8.1305 | 8.4115 | 7.0378 | 6.8569 | 7.049 |
| C1QTNF3 | 5.0113 | 4.666 | 4.8139 | 2.7971 | 3.6614 | 3.2197 |
| TRMT1L | 7.2226 | 6.8926 | 6.737 | 5.7345 | 5.5597 | 6.0376 |
| MSANTD2 | 7.9958 | 8.1209 | 7.9937 | 6.6462 | 6.396 | 6.3063 |
| MPHOSPH8 | 7.1248 | 7.4495 | 7.1136 | 5.9678 | 6.1392 | 5.7641 |
| SYNRG | 7.7835 | 8.2145 | 8.3606 | 6.2298 | 7.1287 | 5.7612 |
| ZNF45 | 6.9248 | 7.1333 | 7.3148 | 6.0632 | 5.9276 | 5.989 |
| IL1R1 | 6.2112 | 6.535 | 6.5129 | 4.1227 | 4.3521 | 4.9581 |
| FAN1 | 6.3098 | 5.7721 | 6.0455 | 4.8679 | 4.4429 | 4.1341 |
| FRY | 5.5217 | 5.0006 | 4.9182 | 3.7877 | 4.0206 | 4.066 |
| CARD8 | 5.3967 | 4.8215 | 5.6734 | 3.9556 | 3.3054 | 3.7684 |
KLF10 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: KLF10
[1] KLF10 as a Tumor Suppressor Gene and Its TGF-beta Signaling.
[2] A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction.
[3] KLF10 Deficiency in CD4(+) T Cells Triggers Obesity, Insulin Resistance, and Fatty Liver.
[4] KLF10 is a modulatory factor of chondrocyte hypertrophy in developing skeleton.
[6] KLF10 Gene Expression Modulates Fibrosis in Dystrophic Skeletal Muscle.
[8] KLF10 transcription factor regulates hepatic glucose metabolism in mice.
[9] Deletion of KLF10 Leads to Stress-Induced Liver Fibrosis upon High Sucrose Feeding.
[10] FBW7 targets KLF10 for ubiquitin-dependent degradation.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below