ZNF207 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN357
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: AG-14361
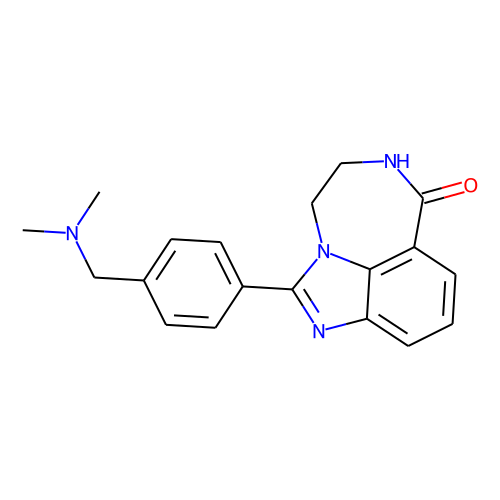
| ID | BRD-K00615600 |
| name | AG-14361 |
| type | Small molecule |
| molecular formula | C19H20N4O |
| molecular weight | 320.4 |
| condition dose | 20 uM |
| condition time | 24 h |
| smiles | CN(C)Cc1ccc(cc1)-c1nc2cccc3C(=O)NCCn1c23 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: ZNF207
Transcription regulatory factors corresponding to this network
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|---|---|
| PTPRH | 5.7523 | 6.0556 | 6.1291 | 6.5659 | 6.8149 | 6.8205 | 6.2156 | 6.5681 | 6.7887 |
| ANGPTL2 | 3.0886 | 3.0999 | 2.6501 | 3.6249 | 3.7858 | 4.1072 | 3.4607 | 4.4099 | 4.4009 |
| PTGDR | 5.0977 | 4.6687 | 4.6048 | 5.5171 | 5.3218 | 5.2818 | 5.5884 | 5.3935 | 5.3406 |
| ZNF557 | 4.8734 | 4.2869 | 4.745 | 5.2066 | 5.4388 | 5.2727 | 5.2197 | 5.6066 | 5.6982 |
| NFKBIA | 9.0254 | 8.843 | 8.9794 | 9.315 | 9.4426 | 9.7004 | 9.6029 | 9.5572 | 9.659 |
| FOSL1 | 5.3843 | 5.6561 | 5.5445 | 6.5623 | 7.0059 | 7.1437 | 7.3573 | 6.5202 | 7.2385 |
| NOTCH1 | 8.1929 | 8.1439 | 8.2649 | 8.9171 | 8.9547 | 8.8239 | 9.1614 | 8.9893 | 8.8888 |
| MAPKAPK2 | 10.0401 | 10.9911 | 10.2339 | 11.0476 | 11.933 | 11.5776 | 11.5796 | 12.3368 | 11.8549 |
| PLCB3 | 8.949 | 9.1303 | 9.2783 | 9.4645 | 9.5411 | 9.8319 | 9.9555 | 9.7376 | 9.7342 |
| DPH2 | 7.5801 | 7.6522 | 7.3556 | 8.6058 | 8.8748 | 8.7889 | 8.8254 | 8.7309 | 8.8987 |
| TRAPPC6A | 9.006 | 8.9612 | 9.8833 | 10.8055 | 10.1191 | 10.0482 | 10.7499 | 10.7004 | 10.7122 |
| CAMSAP2 | 9.2724 | 9.1334 | 9.4881 | 10.883 | 9.9667 | 9.7733 | 9.903 | 10.5673 | 10.316 |
| TOMM34 | 8.7591 | 8.8729 | 8.5405 | 9.9461 | 9.5074 | 9.675 | 9.5378 | 9.0459 | 9.2666 |
| DDIT4 | 9.3253 | 9.2183 | 9.5719 | 10.6702 | 10.3328 | 10.1212 | 10.7004 | 10.7661 | 10.3209 |
| STK25 | 10.209 | 9.9193 | 10.3972 | 11.1525 | 10.8899 | 11.0331 | 11.2952 | 11.2556 | 11.3517 |
| MYC | 10.0831 | 10.114 | 10.0541 | 11.6437 | 10.768 | 11.0583 | 10.9243 | 11.3699 | 11.2665 |
| LRPAP1 | 9.5378 | 9.4865 | 9.4677 | 10.4139 | 10.1413 | 10.6214 | 10.0061 | 10.0082 | 10.13 |
| TCEA2 | 9.2629 | 9.2219 | 9.5719 | 10.266 | 10.2415 | 10.1084 | 10.2757 | 10.5673 | 10.4653 |
| FHL2 | 11.1758 | 11.4815 | 11.1327 | 13.4645 | 12.7054 | 12.8125 | 11.9981 | 12.6668 | 12.5373 |
| TSTA3 | 7.3222 | 7.4661 | 7.2433 | 8.0189 | 8.6863 | 8.6369 | 8.5094 | 8.4536 | 8.3451 |
| GAA | 8.2689 | 8.3107 | 8.1325 | 9.2172 | 9.295 | 9.0696 | 9.5918 | 8.3239 | 9.104 |
| DDB2 | 9.3376 | 9.3587 | 9.5426 | 10.2283 | 9.8493 | 9.6926 | 9.9341 | 10.3933 | 10.0795 |
| NET1 | 9.8602 | 9.4995 | 9.8656 | 10.4139 | 10.5877 | 10.43 | 10.4834 | 10.5055 | 10.4834 |
| P4HTM | 9.5665 | 9.442 | 9.99 | 10.844 | 10.844 | 10.7056 | 11.1555 | 10.7419 | 10.861 |
| GATA3 | 10.6623 | 10.8719 | 10.9042 | 12.0393 | 11.6395 | 11.6987 | 12.0712 | 11.8408 | 11.8648 |
| RFX5 | 7.7239 | 7.7903 | 7.7175 | 8.9065 | 9.0851 | 8.9943 | 9.2533 | 8.9175 | 8.8626 |
| EDEM1 | 7.6442 | 7.4417 | 7.8317 | 8.3258 | 8.0065 | 8.0602 | 8.3239 | 8.6098 | 8.4734 |
| PPIF | 9.5782 | 10.0242 | 9.3213 | 11.7503 | 10.3259 | 11.5608 | 10.2151 | 10.9548 | 10.7579 |
| DALRD3 | 7.5829 | 7.5949 | 7.7316 | 8.9122 | 8.5634 | 8.4292 | 8.1019 | 8.3136 | 8.8239 |
| BTG2 | 4.1642 | 5.0638 | 6.2157 | 7.5512 | 8.1506 | 6.3777 | 7.8499 | 8.4063 | 7.7125 |
| ESRRA | 8.8379 | 9.1846 | 9.0445 | 9.4709 | 9.842 | 9.9009 | 9.4394 | 9.6809 | 9.7838 |
| EIF2S1 | 9.0783 | 9.3867 | 9.3539 | 10.2333 | 10.2137 | 10.274 | 10.0276 | 10.1399 | 9.9153 |
| CHD4 | 8.1532 | 8.174 | 8.1979 | 8.6118 | 8.674 | 8.8102 | 9.1774 | 9.6228 | 9.3581 |
| ITGA3 | 7.8517 | 7.3569 | 7.9377 | 8.5583 | 8.128 | 8.9111 | 8.5818 | 8.9392 | 8.7879 |
| PYGB | 10.4451 | 10.3778 | 10.5367 | 11.5796 | 11.4795 | 11.0998 | 11.3804 | 11.5502 | 10.8021 |
| KRT18 | 11.4458 | 11.9708 | 11.8802 | 13.9046 | 14.5457 | 14.5457 | 12.8412 | 13.7847 | 13.1304 |
| JOSD1 | 7.4316 | 6.8791 | 7.3823 | 7.933 | 7.8319 | 7.8764 | 7.5349 | 8.2283 | 8.0611 |
| UBXN1 | 9.427 | 9.3871 | 9.3042 | 10.1404 | 10.1425 | 10.0401 | 9.6777 | 10.2027 | 10.1811 |
| IMPDH2 | 9.5066 | 9.243 | 9.6717 | 10.305 | 10.8549 | 10.0095 | 10.0891 | 10.4578 | 11.1535 |
| GNL2 | 9.0895 | 8.736 | 9.1327 | 9.6207 | 10.1965 | 10.1241 | 9.5228 | 10.534 | 10.2808 |
| SDC4 | 8.6705 | 9.2968 | 8.7994 | 11.5366 | 11.1466 | 10.2419 | 9.8555 | 11.246 | 10.7255 |
| SPINT1 | 8.2954 | 8.233 | 7.9949 | 8.8148 | 9.4138 | 8.8008 | 8.7671 | 9.2483 | 8.687 |
| CHST15 | 5.3585 | 5.0756 | 5.7155 | 7.6368 | 6.2429 | 6.398 | 7.0701 | 7.6904 | 7.3447 |
| ALDH1A3 | 6.0044 | 5.2229 | 5.7323 | 7.4975 | 7.3606 | 6.8794 | 6.099 | 6.5582 | 6.8916 |
| PDLIM7 | 7.4092 | 7.9713 | 7.7122 | 9.0222 | 9.1929 | 8.1049 | 8.7057 | 8.6632 | 8.5748 |
| IL32 | 6.4374 | 5.9915 | 5.9363 | 7.8439 | 8.4595 | 7.4748 | 7.0566 | 7.9946 | 8.4409 |
| SLC31A2 | 4.6299 | 5.3686 | 4.6833 | 5.7411 | 5.8721 | 5.773 | 5.5877 | 5.8905 | 5.7439 |
| ATP7B | 6.8393 | 6.2741 | 7.0407 | 7.2959 | 7.427 | 7.5821 | 7.5494 | 8.038 | 7.652 |
| RAMP1 | 7.8085 | 7.1154 | 8.1959 | 8.3158 | 9.938 | 9.0364 | 8.6412 | 9.0445 | 9.5949 |
| PIGF | 10.1084 | 9.524 | 9.6672 | 11.0221 | 10.735 | 10.3864 | 10.5233 | 10.6985 | 10.4232 |
| DNAJC4 | 7.9584 | 8.0345 | 7.5963 | 8.4359 | 8.726 | 8.7386 | 8.327 | 8.8851 | 8.5613 |
| ISLR | 5.0516 | 5.1656 | 4.8937 | 7.3749 | 6.2342 | 5.879 | 6.0677 | 7.0477 | 7.7224 |
| ACTN1 | 11.216 | 11.1836 | 11.3517 | 12.9953 | 12.5567 | 11.6672 | 12.1819 | 12.7015 | 12.0577 |
| NDUFV1 | 10.7085 | 11.0904 | 10.9889 | 11.9233 | 11.5174 | 11.5354 | 11.5672 | 12.3258 | 11.6284 |
| KRT7 | 9.9333 | 9.2246 | 8.4095 | 11.4297 | 12.3294 | 12.1819 | 9.7679 | 11.174 | 11.3793 |
| PHLDA2 | 11.7781 | 10.4433 | 10.9415 | 14.1976 | 13.3887 | 12.3611 | 12.1873 | 12.5001 | 13.1782 |
| LAMA5 | 7.0131 | 8.0846 | 7.5869 | 8.6293 | 8.5352 | 8.3221 | 8.2427 | 8.5849 | 8.1465 |
| PLAUR | 3.1937 | 2.9847 | 3.8377 | 4.145 | 4.2403 | 4.3055 | 4.5652 | 4.2843 | 4.1149 |
| ATP6V0D1 | 10.4381 | 11.382 | 10.5704 | 12.0577 | 11.2092 | 11.4999 | 11.8266 | 11.7047 | 12.0525 |
| AP2A2 | 8.3908 | 8.1964 | 8.4751 | 8.8892 | 9.2048 | 8.8581 | 9.5858 | 9.0675 | 9.5378 |
| NEK9 | 5.3969 | 4.9209 | 5.5072 | 6.0635 | 6.5826 | 5.9336 | 5.9086 | 5.9333 | 5.951 |
| SCRIB | 10.0772 | 10.5005 | 9.6535 | 11.3237 | 11.7176 | 11.9981 | 10.8321 | 11.7143 | 11.2538 |
| DENND5A | 7.2348 | 6.9395 | 7.1943 | 8.3865 | 8.0716 | 8.5758 | 7.6012 | 7.7933 | 7.6542 |
| TNPO1 | 9.0664 | 9.4639 | 9.2134 | 9.8786 | 10.07 | 10.0068 | 9.5713 | 9.976 | 10.1511 |
| ZNF3 | 7.9401 | 8.1064 | 8.1348 | 8.8783 | 8.6918 | 8.8318 | 8.721 | 9.0053 | 9.5399 |
| PACS2 | 7.3245 | 6.9621 | 7.0842 | 8.0576 | 7.9968 | 7.5427 | 8.0677 | 8.2878 | 8.0053 |
| GATAD1 | 6.5061 | 6.8819 | 6.9767 | 7.5135 | 7.8967 | 7.4063 | 7.9468 | 7.2543 | 7.5729 |
| ARHGEF18 | 7.3193 | 7.4627 | 7.9966 | 8.4521 | 8.5688 | 8.6846 | 8.7201 | 8.8197 | 9.3026 |
| ZFYVE26 | 5.5636 | 5.7243 | 5.9791 | 6.2109 | 6.2752 | 6.1123 | 6.424 | 6.509 | 6.6593 |
| HSPA6 | 6.1363 | 5.3713 | 4.3881 | 6.9151 | 7.3556 | 7.059 | 6.8242 | 6.6681 | 7.3795 |
| NOP16 | 11.8874 | 11.3632 | 11.8471 | 12.5094 | 12.5277 | 12.4349 | 12.5024 | 12.9241 | 12.4897 |
| WDR43 | 9.4692 | 10.292 | 10.0417 | 11.4415 | 11.0531 | 10.9558 | 10.6276 | 10.514 | 10.6679 |
| ZKSCAN1 | 6.008 | 6.0133 | 5.2158 | 7.167 | 7.5344 | 6.7348 | 6.7773 | 7.4074 | 6.947 |
| NBPF10 | 6.0471 | 5.8352 | 5.7828 | 6.5731 | 6.4762 | 6.5471 | 6.951 | 7.3594 | 6.8011 |
| BCL6 | 5.7402 | 5.1646 | 5.1159 | 6.8759 | 6.7759 | 6.0384 | 6.2007 | 6.7137 | 6.7135 |
| SFXN3 | 6.7259 | 6.8983 | 6.879 | 7.26 | 7.7346 | 7.4254 | 7.8698 | 7.9964 | 7.6986 |
| GSTK1 | 11.0597 | 10.5655 | 10.9806 | 12.5277 | 11.9501 | 12.1088 | 11.8218 | 12.1306 | 11.8771 |
| FAM160B2 | 7.6497 | 8.0476 | 7.5087 | 8.7829 | 8.4525 | 8.1165 | 8.6069 | 8.635 | 8.3435 |
| MICAL1 | 8.5389 | 8.2743 | 8.7432 | 9.6348 | 9.3057 | 9.6098 | 9.5273 | 9.9037 | 10.1569 |
| PRKRIP1 | 7.6769 | 7.8305 | 8.1617 | 8.1691 | 8.5439 | 8.4625 | 8.6378 | 8.7558 | 8.5785 |
| DNAAF5 | 8.5951 | 8.8076 | 8.6532 | 9.2396 | 9.4872 | 9.3376 | 9.1399 | 9.6311 | 9.8029 |
| IER5 | 10.3547 | 9.6338 | 9.5998 | 11.1602 | 10.9429 | 12.0448 | 11.246 | 11.9061 | 11.4942 |
| PUS7 | 9.2027 | 9.386 | 9.982 | 10.4823 | 11.0956 | 10.7192 | 10.5541 | 10.9359 | 10.8589 |
| FBXL6 | 7.5583 | 7.7874 | 7.3482 | 8.8049 | 8.6486 | 8.3422 | 7.8726 | 8.4701 | 8.6154 |
| SPHK1 | 6.3531 | 5.9791 | 5.8615 | 7.5333 | 6.8879 | 6.4419 | 6.7478 | 7.3954 | 7.0472 |
| MICALL2 | 8.5274 | 8.6409 | 8.3113 | 9.5537 | 9.4611 | 9.3678 | 8.9083 | 9.7523 | 9.4286 |
| ZNF267 | 5.3426 | 5.4511 | 5.9174 | 6.5015 | 6.5839 | 6.2204 | 6.2098 | 6.1763 | 6.327 |
| ZMAT3 | 4.6549 | 4.8531 | 4.6143 | 5.7097 | 5.4833 | 5.5362 | 5.3131 | 6.2767 | 5.6587 |
| ENGASE | 7.0833 | 7.1275 | 7.1572 | 7.5948 | 7.8182 | 8.7215 | 7.9939 | 8.3447 | 7.8564 |
| SLC25A32 | 11.3347 | 11.319 | 11.7559 | 12.8316 | 12.3384 | 11.8266 | 12.1163 | 12.1219 | 12.6432 |
| SCLY | 8.4261 | 8.1406 | 8.0119 | 9.2091 | 9.3761 | 9.0159 | 8.9777 | 9.0424 | 9.314 |
| RRN3 | 9.4023 | 9.0598 | 9.5033 | 9.8602 | 10.2057 | 10.2267 | 10.0736 | 10.4715 | 9.9216 |
| PPP1R10 | 6.6357 | 6.613 | 6.984 | 7.1898 | 7.2464 | 7.4159 | 7.7822 | 7.3086 | 7.4358 |
| FCGR2A | 4.4172 | 3.4087 | 4.1237 | 4.9995 | 4.749 | 5.6502 | 5.4488 | 5.4596 | 5.8751 |
| AMPD1 | 4.2704 | 4.0715 | 4.7459 | 5.0588 | 4.8882 | 4.6932 | 5.1084 | 5.255 | 5.266 |
| HBZ | 4.4025 | 4.0624 | 4.9506 | 5.3125 | 5.2494 | 4.9255 | 5.1769 | 5.6517 | 5.3126 |
ZNF207 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network