NIPBL transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN4068
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: wortmannin
condition dose: 10 nM
condition time: 6 h
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: NIPBL
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
PI4KA 
|
hsa04070 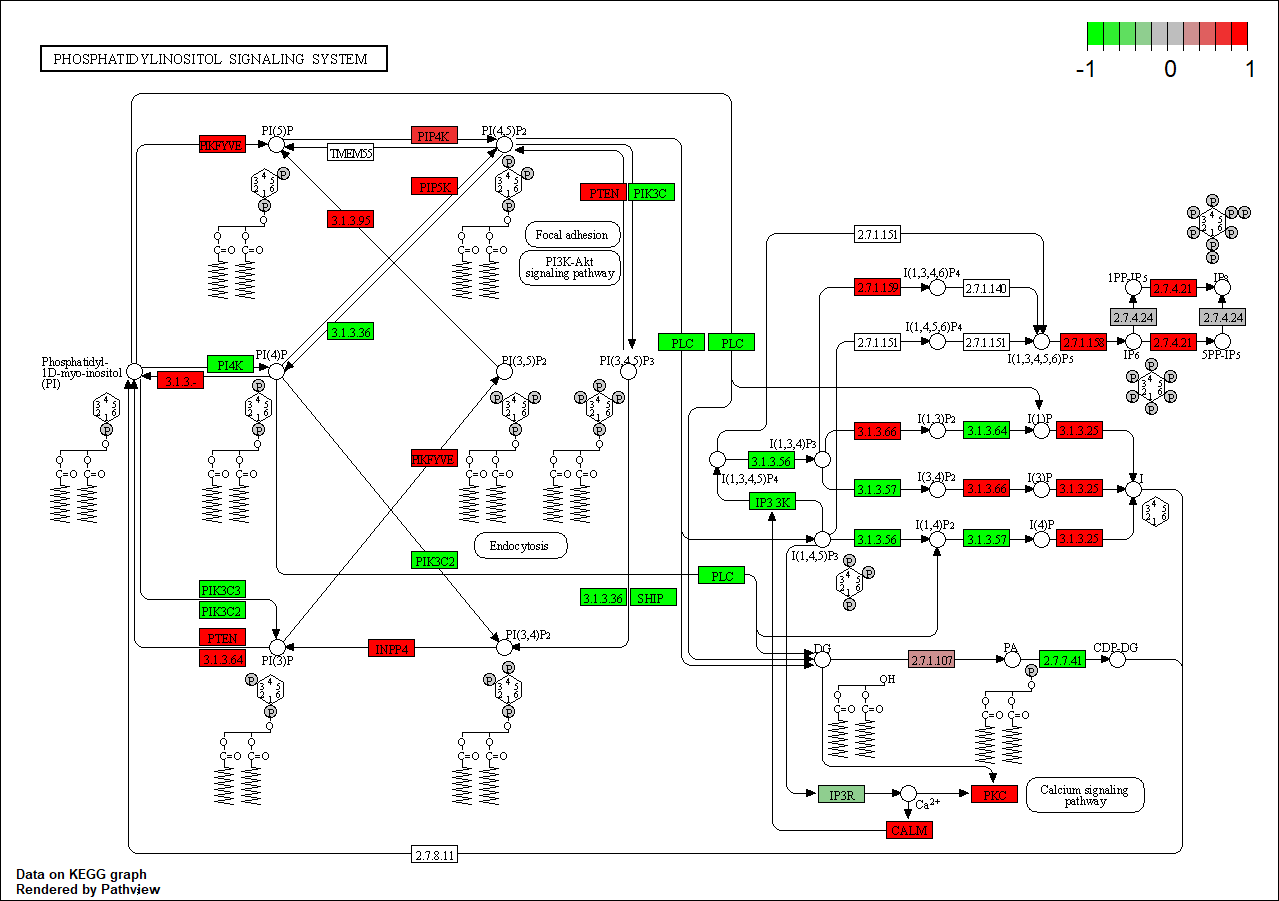
|
|
PI4KB 
|
hsa00562 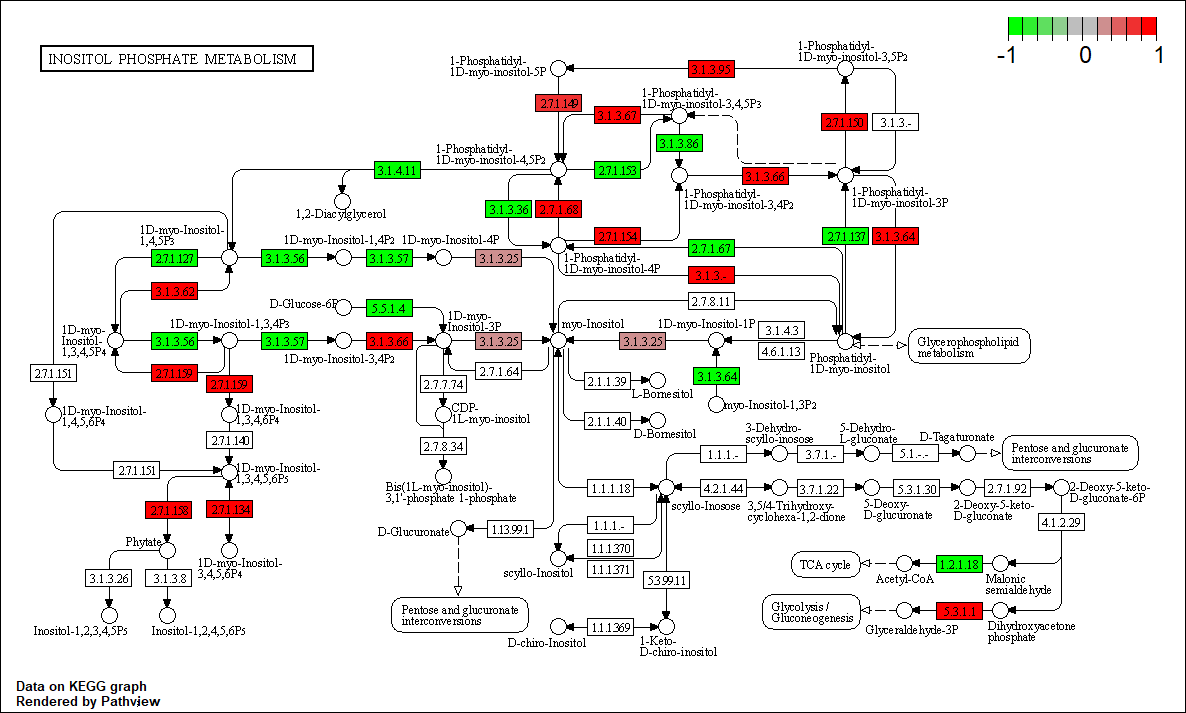
|
|
PIK3CA 
|
hsa04140 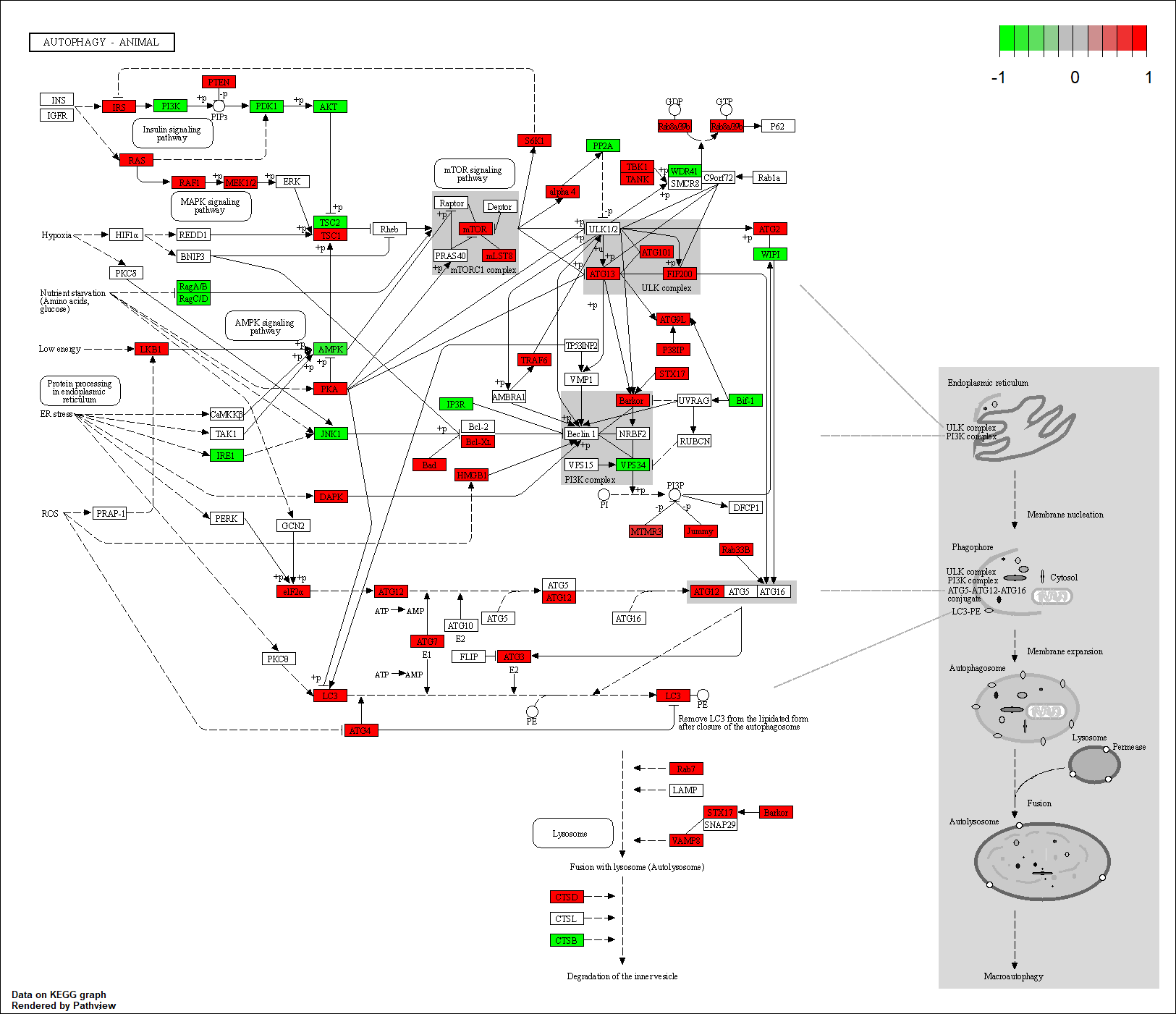
|
|
PIK3CD 
|
hsa00562 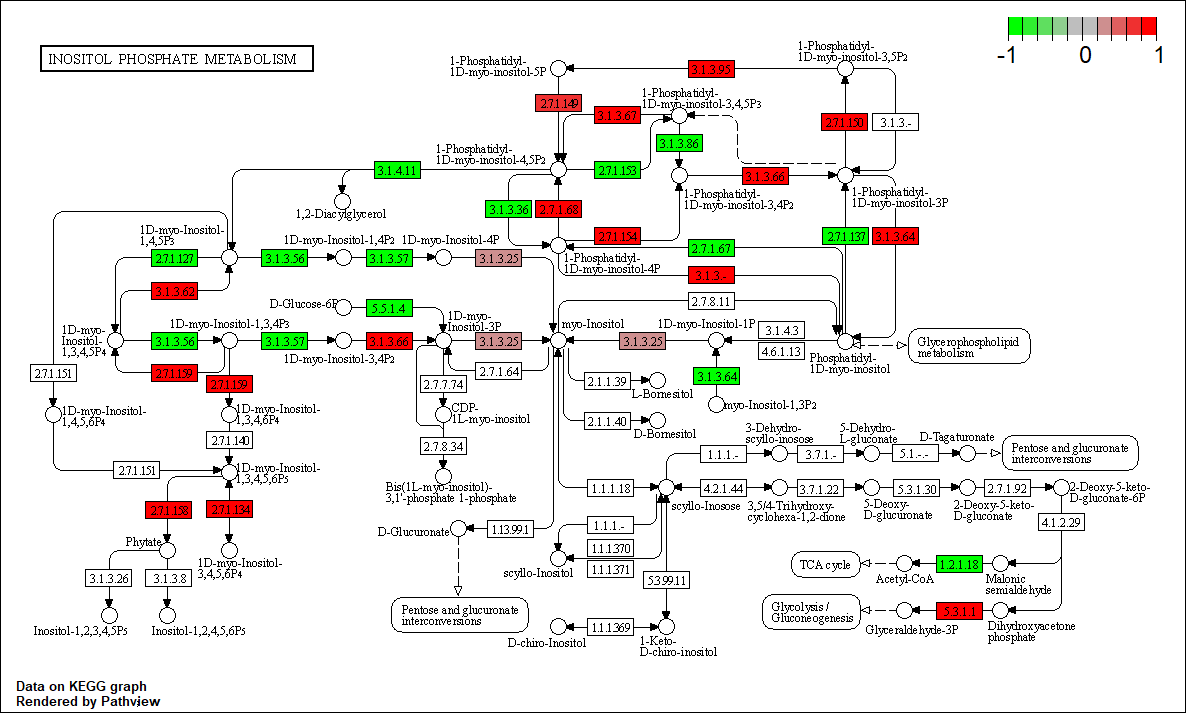
|
|
PIK3CG 
|
hsa04261 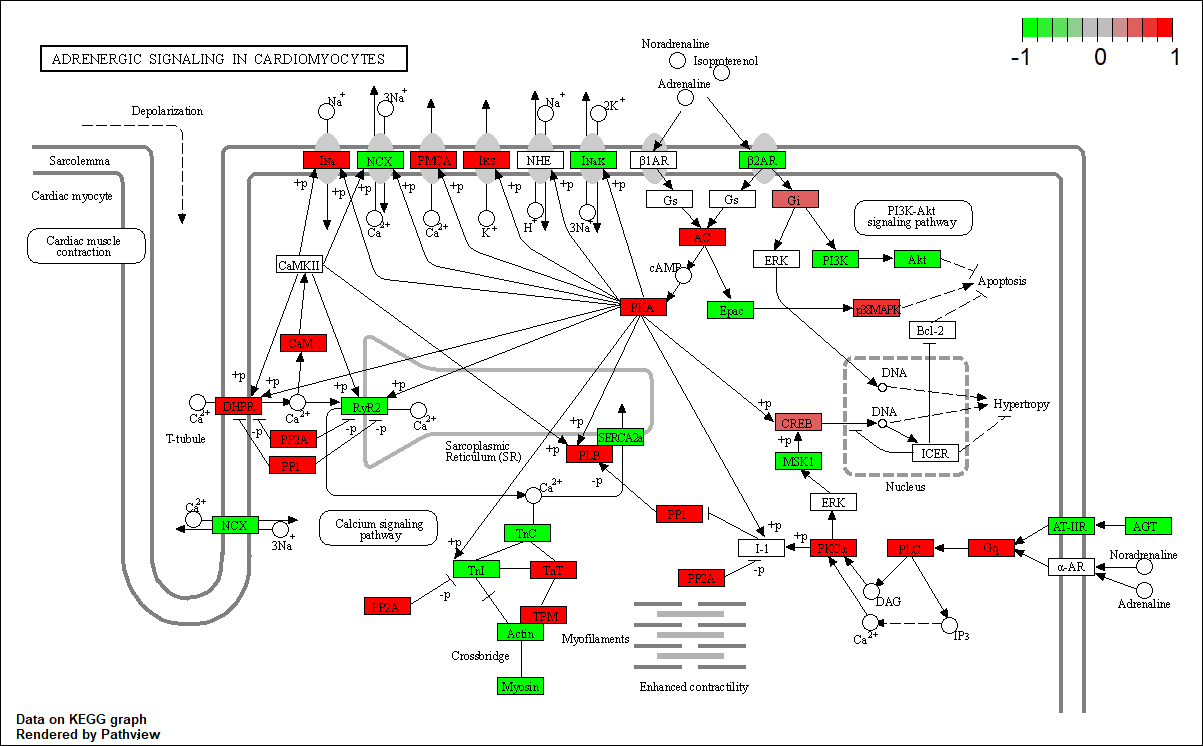
|
|
PIK3R1 
|
hsa04668 
|
|
PLK1 
|
hsa04914 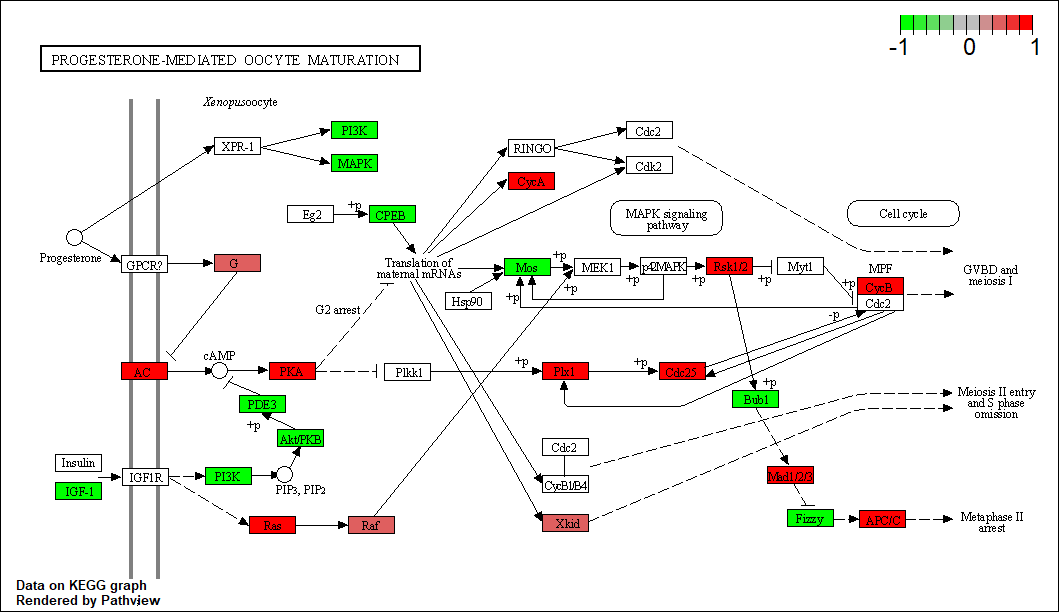
|
|
PRKDC 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition |
|---|---|---|---|---|
| CD3D | 4.8637 | 5.0318 | 4.7517 | 0.4563 |
| IRX4 | 6.4148 | 6.5229 | 6.5101 | 3.0823 |
| ATF1 | 8.1642 | 8.3402 | 8.4955 | 1.8883 |
| BRCA1 | 7.1737 | 7.5127 | 7.5826 | 2.7292 |
| HSPA8 | 11.7582 | 11.6119 | 11.276 | 4.8889 |
| CYCS | 9.9471 | 9.8721 | 10.1989 | 4.9458 |
| NMT1 | 8.9174 | 8.7231 | 8.8969 | 2.8522 |
| HDAC2 | 10.5367 | 10.357 | 10.6721 | 6.6422 |
| MBTPS1 | 8.4156 | 8.5289 | 8.5375 | 5.1026 |
| LPGAT1 | 8.2517 | 8.8396 | 8.3057 | 2.5961 |
| GLRX | 6.6298 | 6.9097 | 6.9717 | 2.7292 |
| POLG2 | 9.0795 | 9.405 | 9.1498 | 2.5114 |
| TRAPPC3 | 11.2085 | 11.6502 | 11.6595 | 6.3905 |
| RBM34 | 10.0436 | 10.0735 | 10.3549 | 4.1332 |
| RAD51C | 10.9929 | 11.1154 | 11.4979 | 4.7783 |
| YME1L1 | 9.8342 | 9.8824 | 9.9216 | 5.0522 |
| POLR2I | 12.0611 | 11.9481 | 11.3525 | 5.2229 |
| HAT1 | 9.5662 | 9.3915 | 9.3444 | 2.6424 |
| ENOPH1 | 11.355 | 11.4459 | 12.1739 | 5.0522 |
| JMJD6 | 12.0705 | 12.1635 | 12.3343 | 5.0522 |
| NOSIP | 10.1448 | 10.2605 | 9.6596 | 4.6662 |
| NUSAP1 | 10.2373 | 10.2373 | 10.4405 | 4.2235 |
| SCAND1 | 12.1345 | 11.9408 | 11.6172 | 7.2008 |
| FIS1 | 11.0787 | 11.4881 | 11.1144 | 5.1464 |
| SPRED2 | 10.0247 | 10.1426 | 10.2687 | 6.4871 |
| TXNDC9 | 9.992 | 9.7915 | 9.7748 | 2.3649 |
| ZMYM2 | 11.4247 | 11.3864 | 11.3018 | 6.4871 |
| NPEPL1 | 11.5998 | 11.6808 | 11.2887 | 4.6662 |
| HOMER2 | 9.2239 | 9.1779 | 9.4537 | 4.3624 |
| CLTB | 11.4392 | 11.6887 | 11.9892 | 5.8642 |
| CCDC86 | 11.421 | 11.3745 | 11.4428 | 7.4796 |
| IFRD2 | 12.4639 | 11.9715 | 12.3092 | 5.2663 |
| ISOC1 | 11.2691 | 10.7298 | 11.3948 | 5.3995 |
| MACF1 | 9.5662 | 9.6038 | 9.1498 | 4.4775 |
| PMM2 | 13.0779 | 12.6645 | 12.8659 | 7.8169 |
| PRR7 | 10.5279 | 10.7465 | 10.0596 | 4.8234 |
| ITGAE | 10.6795 | 10.7755 | 10.6894 | 4.8889 |
| NUP85 | 11.4836 | 11.8267 | 11.2253 | 5.3995 |
| MRPS16 | 11.9156 | 11.8661 | 11.6904 | 5.6981 |
| NOL3 | 10.7782 | 10.9153 | 11.0521 | 6.7652 |
| NR2F6 | 11.1782 | 11.4377 | 10.8404 | 5.3995 |
| ACAT2 | 10.0897 | 10.1836 | 9.8514 | 6.0553 |
| CHMP4A | 10.587 | 10.6959 | 10.5954 | 2.9588 |
| FBXO21 | 11.3826 | 11.1306 | 11.3213 | 6.3407 |
| GATA3 | 11.287 | 11.4543 | 11.2684 | 4.6662 |
| NVL | 11.3462 | 10.8094 | 11.0773 | 5.9584 |
| ATF5 | 11.5012 | 11.9132 | 11.7491 | 6.1839 |
| UFM1 | 10.372 | 10.2204 | 10.1556 | 5.9584 |
| PTPRK | 11.2483 | 10.7596 | 11.1096 | 5.9584 |
| RAB31 | 9.1935 | 9.3104 | 9.0927 | 5.2663 |
| RFC5 | 10.5589 | 10.7987 | 10.431 | 6.6193 |
| CEP55 | 9.3531 | 9.2222 | 9.402 | 5.2037 |
| ZNF146 | 10.8229 | 11.1415 | 11.1746 | 6.3596 |
| TAF10 | 10.5074 | 10.3472 | 9.99 | 5.3425 |
| BZW1 | 11.4234 | 11.8204 | 11.6439 | 5.1621 |
| ATF4 | 11.8333 | 11.8965 | 11.562 | 5.6711 |
| STRAP | 12.0093 | 11.7882 | 12.2146 | 7.3049 |
| NOP56 | 13.9025 | 13.7838 | 13.8878 | 10.3321 |
| ACSL3 | 12.4708 | 12.6755 | 12.8689 | 7.9803 |
| UBXN1 | 9.8336 | 10.2948 | 9.9779 | 5.4066 |
| RRM2 | 12.9244 | 12.8569 | 12.6822 | 7.455 |
| WDR82 | 9.8409 | 10.0932 | 10.1129 | 6.2007 |
| GGPS1 | 9.1329 | 8.8778 | 9.177 | 4.658 |
| MCRS1 | 10.1072 | 10.2981 | 9.9037 | 5.746 |
| ARL3 | 8.551 | 8.6138 | 8.3774 | 4.488 |
| TOB1 | 11.1902 | 11.5114 | 11.0972 | 4.9075 |
| LSM4 | 11.5058 | 11.8864 | 11.7478 | 7.675 |
| RNASEH2A | 10.8268 | 10.9857 | 10.6196 | 5.1192 |
| BCAS2 | 12.4187 | 11.9168 | 12.6524 | 5.5718 |
| DTYMK | 10.8605 | 10.8491 | 10.8312 | 5.2993 |
| RPP30 | 8.2083 | 8.1569 | 8.2585 | 4.6258 |
| PNO1 | 9.205 | 9.1662 | 9.0364 | 5.3386 |
| MYL6B | 10.4447 | 10.3188 | 10.465 | 5.9459 |
| CD83 | 6.5756 | 6.6123 | 6.8932 | 2.6537 |
| SEC14L2 | 7.5963 | 7.3333 | 7.1627 | 2.6861 |
| FEN1 | 13.3575 | 13.3788 | 13.6934 | 8.938 |
| TTK | 8.9037 | 8.8832 | 9.0052 | 5.4825 |
| NUFIP1 | 8.8249 | 8.7094 | 8.5933 | 5.0605 |
| DNAJC8 | 10.9004 | 10.889 | 10.7502 | 7.0317 |
| BLM | 5.7464 | 6.0193 | 6.0071 | 1.7351 |
| GALNS | 9.1662 | 9.1199 | 8.8621 | 5.1807 |
| TOP1 | 11.1378 | 11.0638 | 11.1103 | 7.1978 |
| NXF1 | 9.9199 | 9.5672 | 9.7242 | 5.4766 |
| GABARAPL2 | 9.9785 | 9.448 | 9.6977 | 4.8816 |
| NHP2 | 14.8455 | 14.4145 | 14.6341 | 10.2744 |
| CNPY2 | 11.2309 | 11.1665 | 11.4946 | 6.759 |
| TUBD1 | 11.0594 | 11.5177 | 11.3399 | 6.3173 |
| CEBPB | 15 | 15 | 15 | 7.0946 |
| PTPN11 | 10.1311 | 10.3947 | 10.5454 | 5.872 |
| RND3 | 12.8396 | 12.7582 | 13.715 | 4.19 |
| OIP5 | 9.5882 | 9.7666 | 9.5082 | 5.3657 |
| APRT | 13.6446 | 13.65 | 13.2432 | 8.5065 |
| NOP16 | 14.1883 | 14.2085 | 14.2971 | 9.1472 |
| GGCT | 12.536 | 12.0581 | 12.7844 | 6.1896 |
| WBP11 | 10.9794 | 10.8373 | 11.32 | 6.2729 |
| RNF130 | 10.3608 | 9.8631 | 10.4522 | 3.2882 |
| PMEPA1 | 7.7926 | 7.8951 | 7.7164 | 1.9806 |
| VKORC1 | 11.2513 | 11.0939 | 10.6375 | 5.8474 |
| CFAP20 | 9.2322 | 9.7617 | 9.8484 | 4.0317 |
| SAP30BP | 10.924 | 11.0951 | 11.1435 | 6.2964 |
| CXCL14 | 11.6226 | 11.4979 | 11.8864 | 1.7649 |
| PRPF40A | 9.9907 | 9.7182 | 9.7957 | 5.0535 |
| TIMM13 | 11.0521 | 10.925 | 11.2691 | 6.4753 |
| NDUFB4 | 12.3815 | 12.3068 | 12.1385 | 8.4706 |
| NUBP2 | 10.7024 | 10.7782 | 10.773 | 7.4542 |
| DDA1 | 8.8681 | 9.1927 | 9.1261 | 4.6416 |
| ZWILCH | 10.2575 | 9.9792 | 10.5398 | 4.5049 |
| CMC2 | 11.7698 | 11.6985 | 12.0705 | 7.0467 |
| WDR59 | 10.417 | 10.1906 | 10.211 | 6.696 |
| CNIH4 | 11.2338 | 11.0835 | 11.2803 | 5.1945 |
| PUS7 | 12.1153 | 11.9587 | 12.0753 | 6.4606 |
| NIP7 | 10.8932 | 11.0951 | 10.5761 | 4.1447 |
| ZNF576 | 9.6383 | 9.6355 | 9.3787 | 5.7702 |
| TMEM160 | 10.9153 | 11.3266 | 10.7551 | 5.7708 |
| VANGL1 | 8.9233 | 9.239 | 9.0368 | 4.9573 |
| GTDC1 | 5.4365 | 5.52 | 5.6792 | 1.9721 |
| CSDE1 | 12.3233 | 12.2527 | 12.2674 | 7.8609 |
| MRTO4 | 11.9086 | 11.7312 | 11.6539 | 7.3333 |
| SLC38A2 | 11.6969 | 11.8745 | 12.0219 | 7.4547 |
| PLEKHA5 | 12.4221 | 12.0942 | 12.6341 | 6.3562 |
| BRIP1 | 8.8146 | 8.7577 | 8.918 | 4.6316 |
| BABAM1 | 11.2102 | 11.6636 | 11.343 | 6.1341 |
| WDR74 | 10.3478 | 10.5385 | 10.3184 | 6.6455 |
| GPSM2 | 9.8699 | 9.951 | 10.1145 | 5.8173 |
| GLRX5 | 9.7366 | 9.8929 | 9.7419 | 5.7157 |
| CERS2 | 10.2138 | 10.1634 | 10.6278 | 4.9819 |
| CXADR | 9.4287 | 10.0156 | 9.9393 | 3.3751 |
| TULP3 | 10.7649 | 10.49 | 10.5595 | 5.5255 |
NIPBL transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: NIPBL
[1] Cryo-EM structure of the human cohesin-NIPBL-DNA complex.
[2] Roles of NIPBL in maintenance of genome stability.
[4] BRD4 interacts with NIPBL and BRD4 is mutated in a Cornelia de Lange-like syndrome.
[6] NIPBL: a new player in myeloid cell differentiation.
[7] Cornelia de Lange syndrome.
[9] Cornelia de Lange syndrome: from molecular diagnosis to therapeutic approach.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below