PAF1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN424
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: dorsomorphin
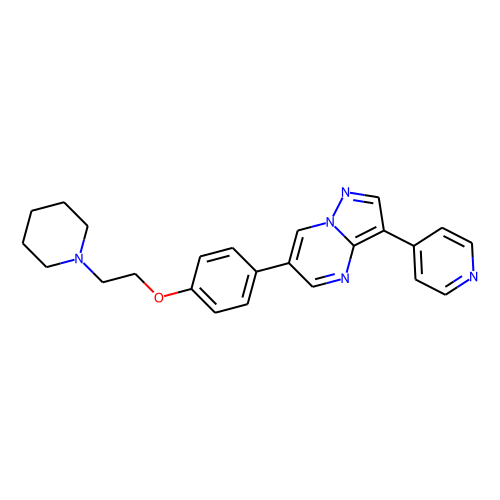
| ID | BRD-K54233340 |
| name | dorsomorphin |
| type | Small molecule |
| molecular formula | C24H25N5O |
| molecular weight | 399.5 |
| condition dose | 10 uM |
| condition time | 24 h |
| smiles | C(CN1CCCCC1)Oc1ccc(cc1)-c1cnc2c(cnn2c1)-c1ccncc1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: PAF1
Transcription regulatory factors corresponding to this network
ARTICLE: dorsomorphin
[1] Zebrafish as tools for drug discovery.
[2] Compound C/Dorsomorphin: Its Use and Misuse as an AMPK Inhibitor.
[3] Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism.
[4] Role of AMP-activated protein kinase in mechanism of metformin action.
[8] Bilobalide Suppresses Adipogenesis in 3T3-L1 Adipocytes via the AMPK Signaling Pathway.
[9] Bone morphogenetic protein receptors and signal transduction.
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
ACVR1 
|
hsa04350 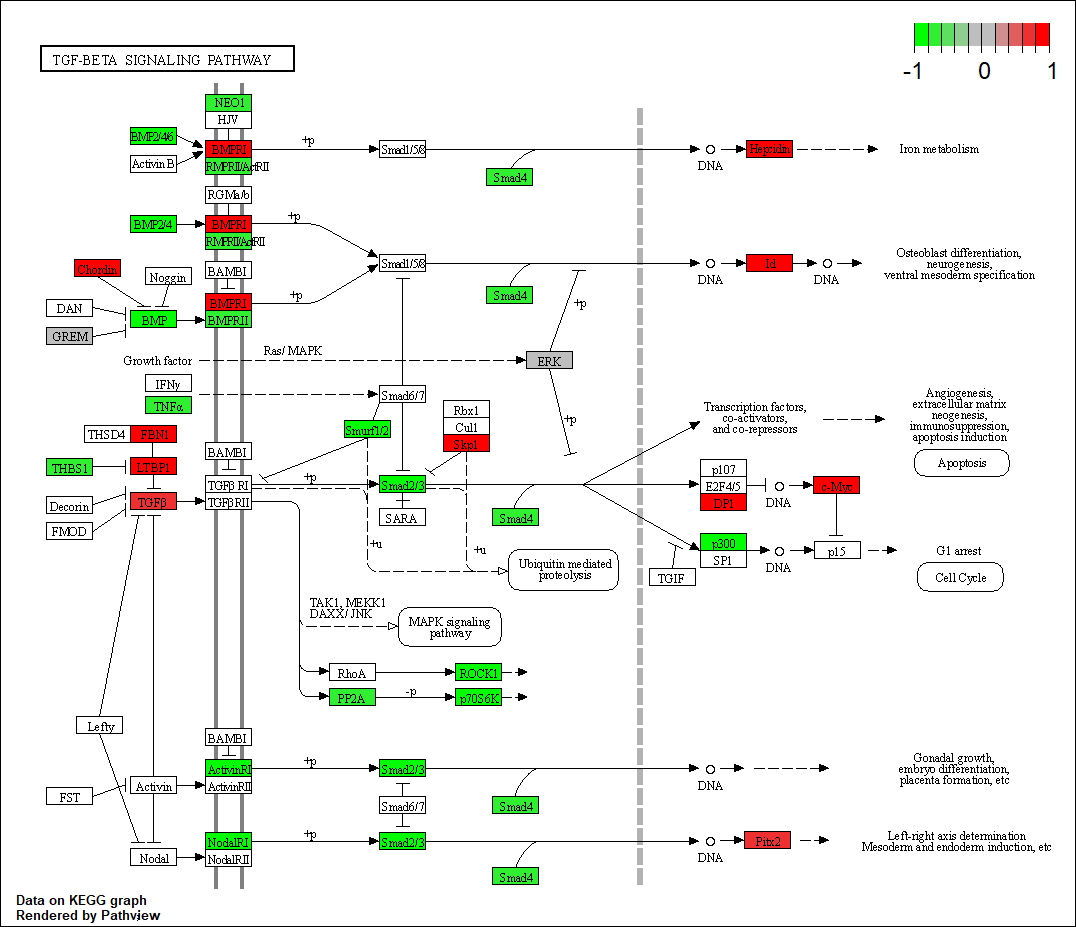
|
|
BMPR1A 
|
hsa04390 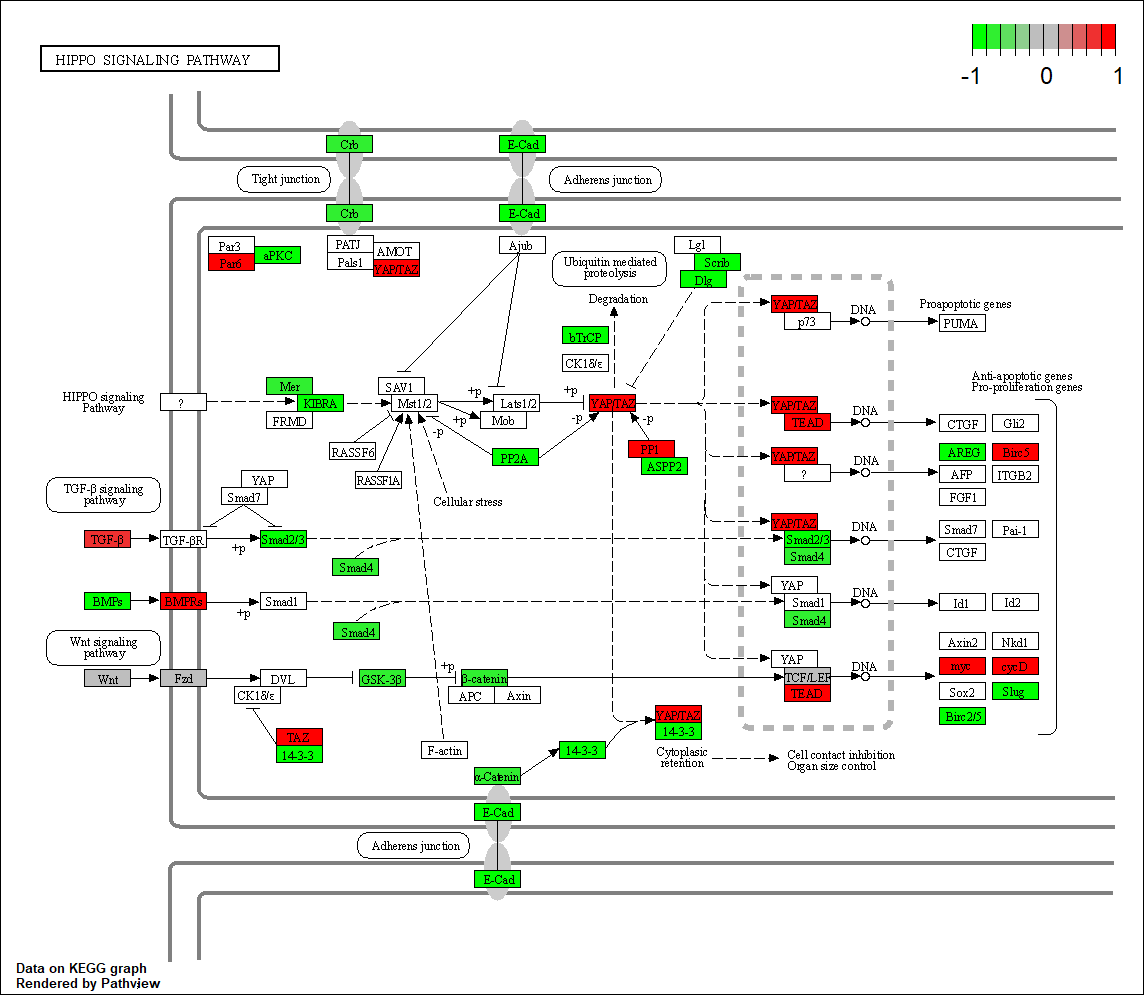
|
|
BMPR1B |
hsa04390 
|
|
EPHA2 
|
hsa04360 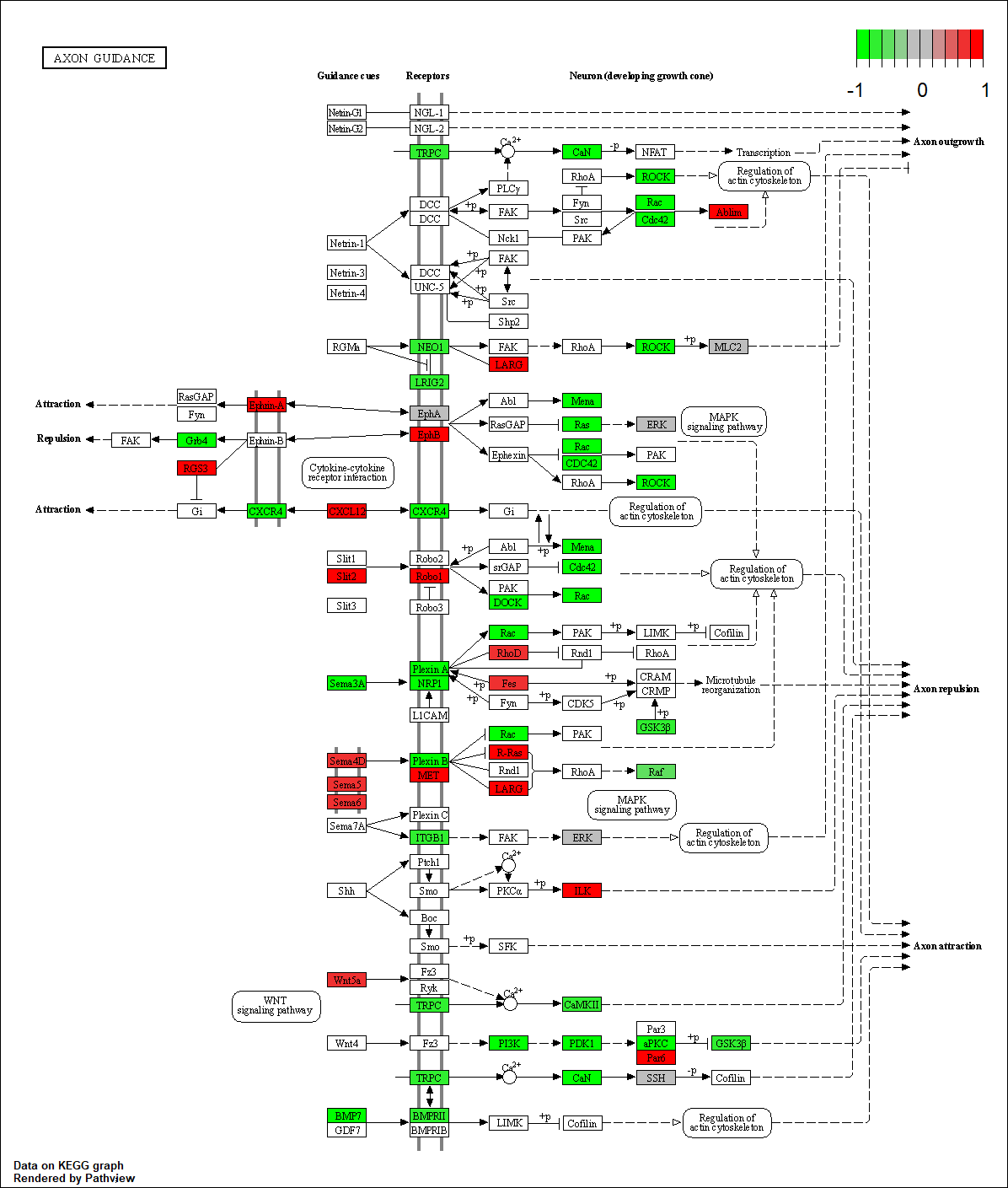
|
|
FKBP1A |
|
|
FLT1 
|
hsa04010 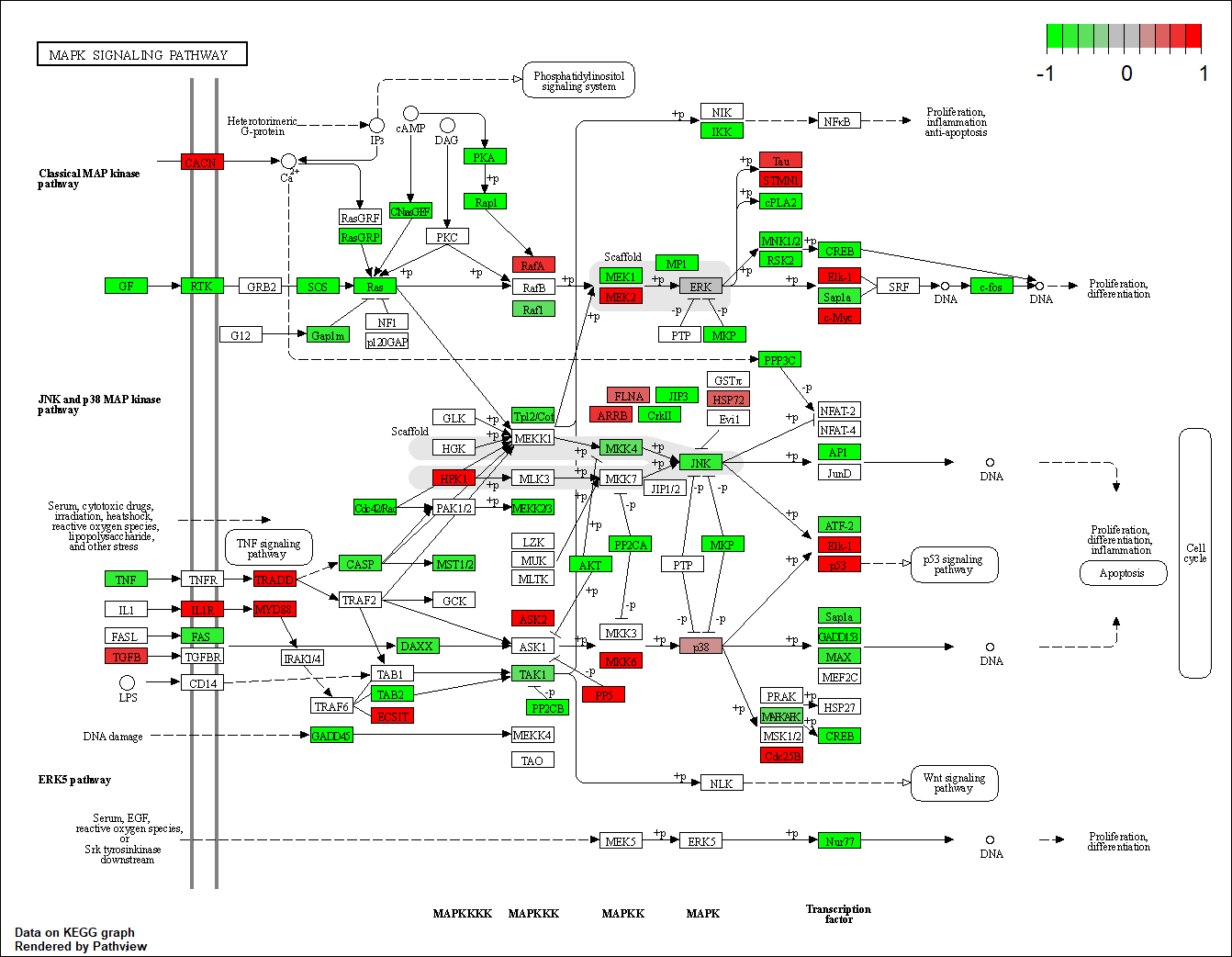
|
|
FLT3 
|
hsa04010 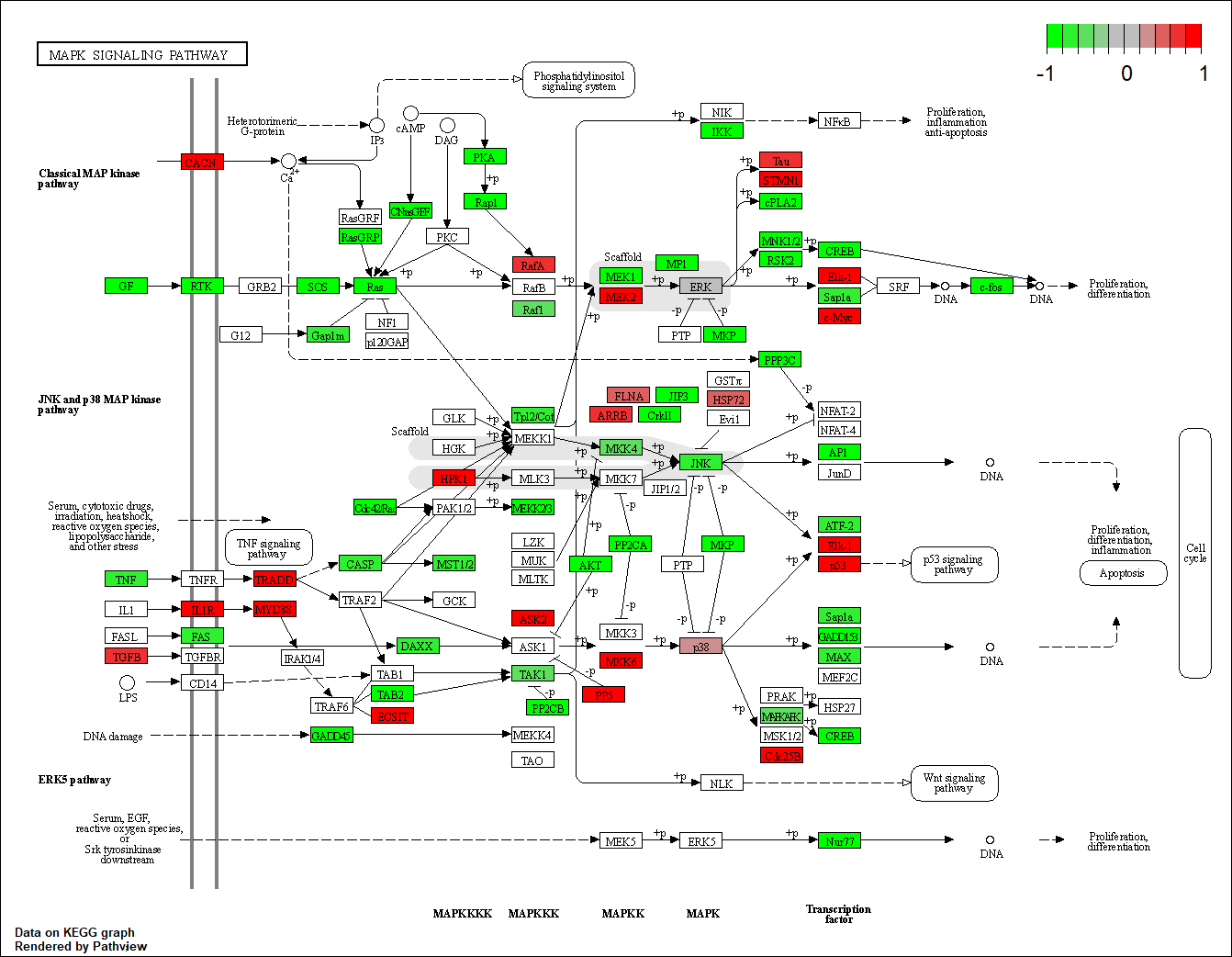
|
|
KDR 
|
hsa04510 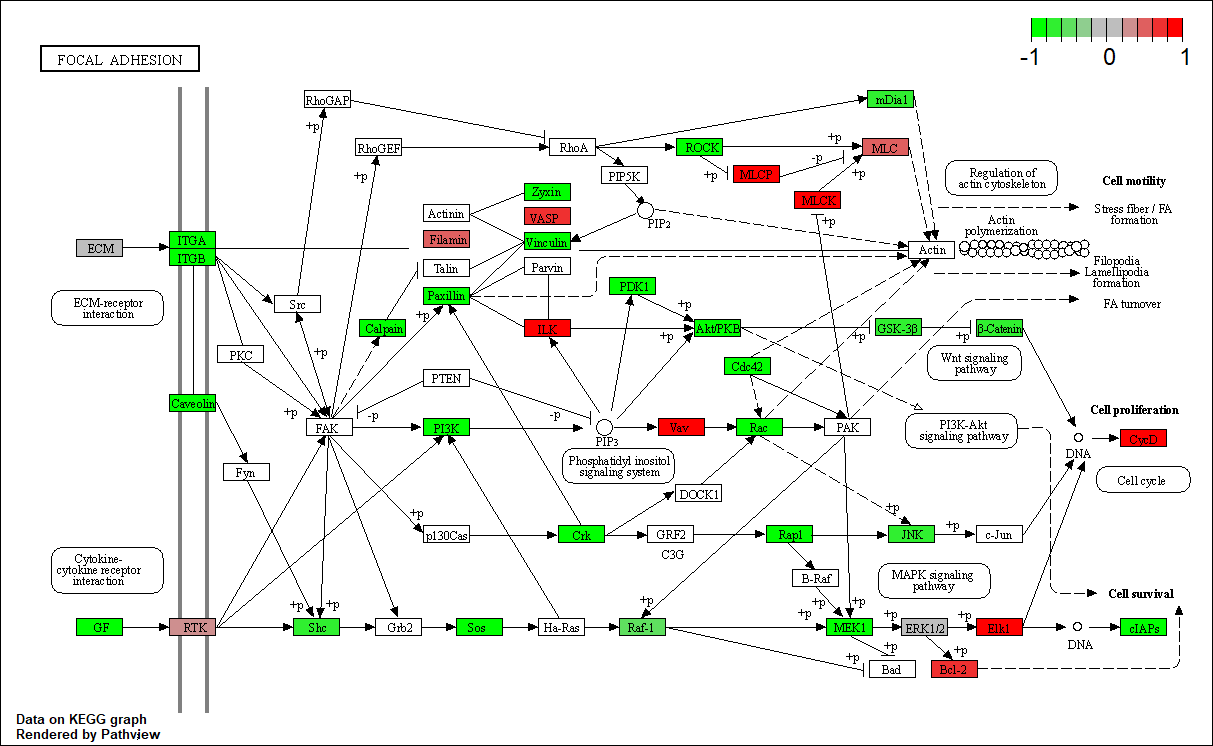
|
|
LCK |
hsa05166 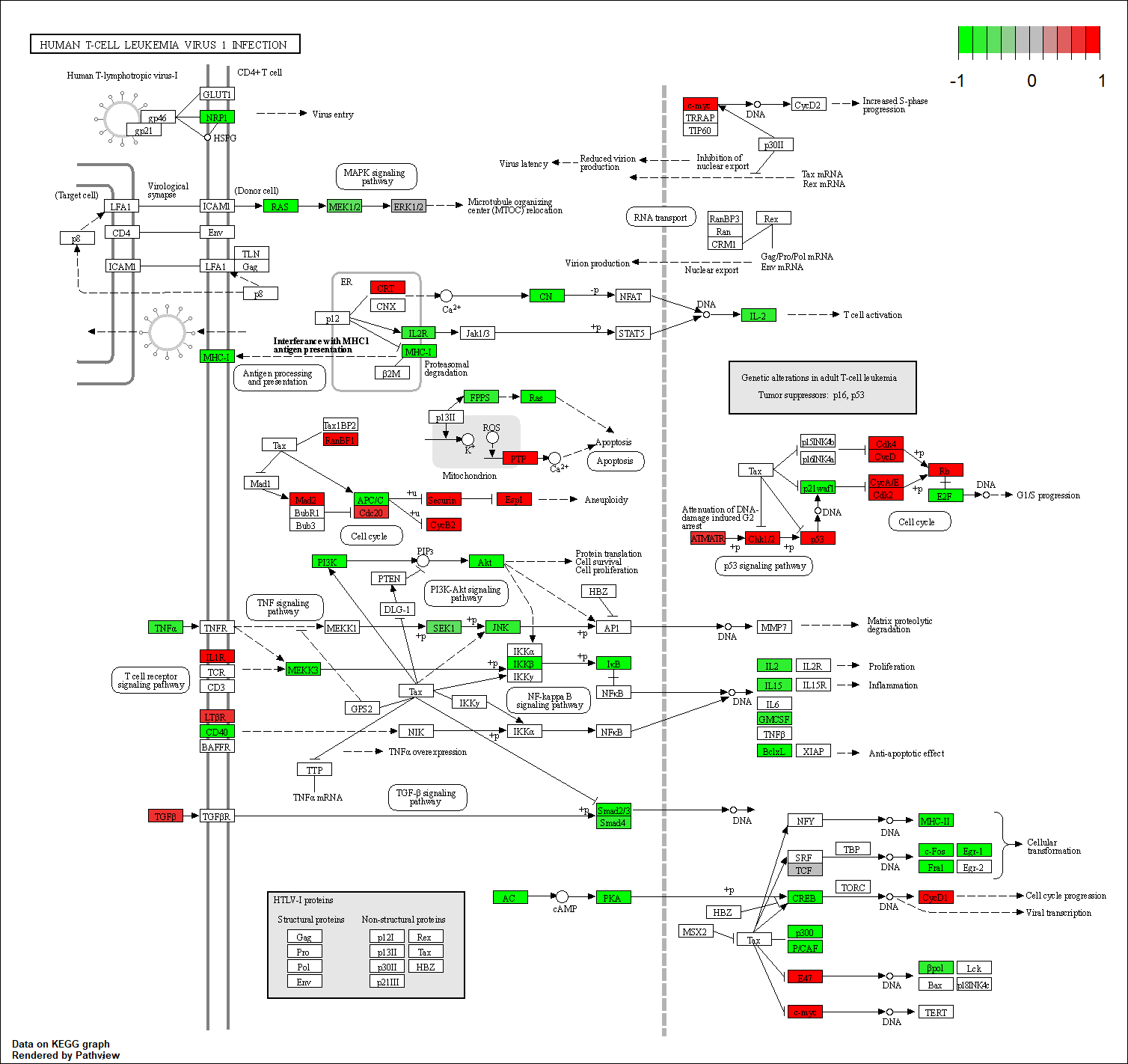
|
|
MKNK1 
|
hsa04910 
|
|
PRKAA1 
|
hsa04932 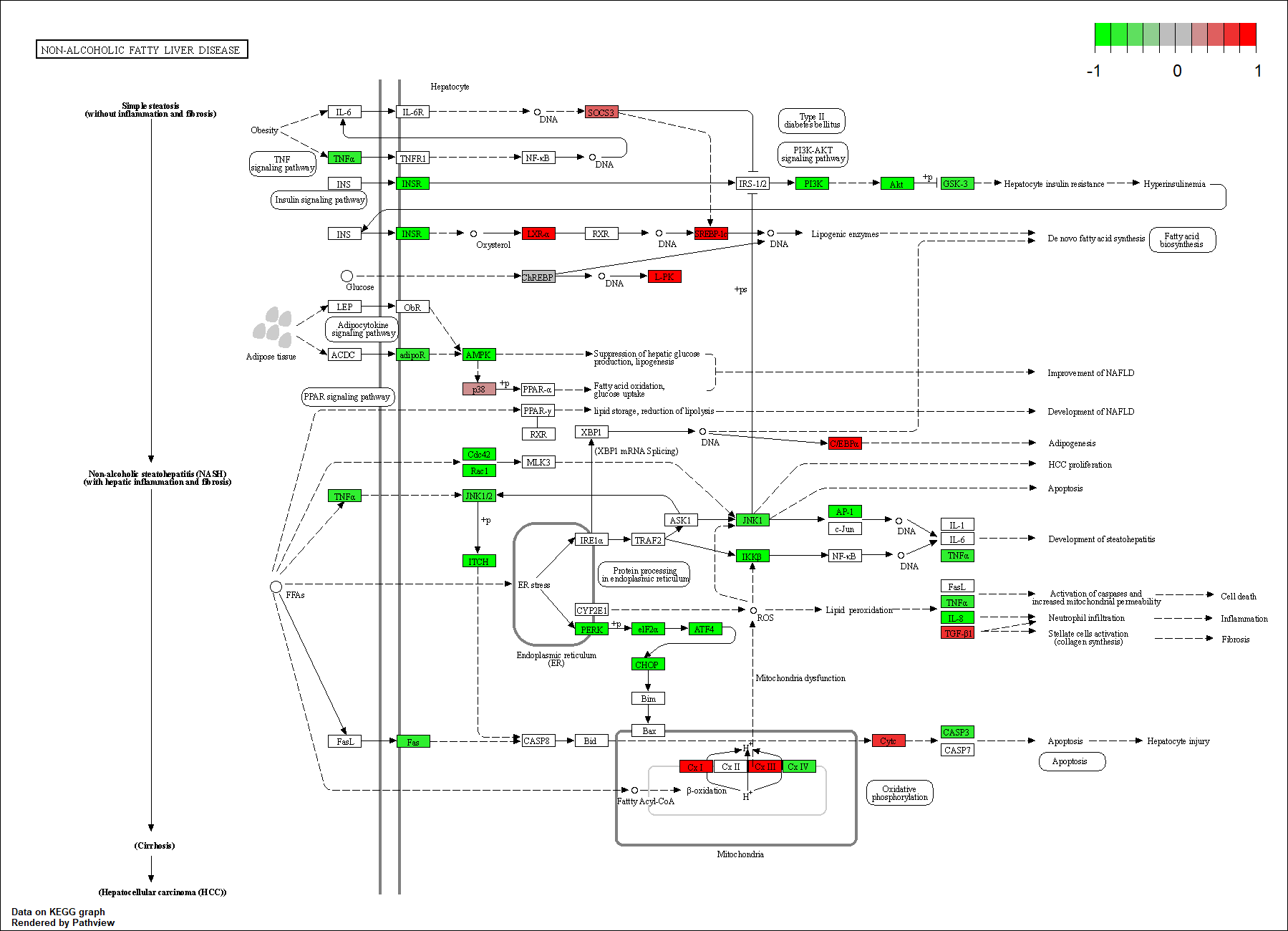
|
|
RPS6KA1 
|
hsa05207 
|
|
SRC 
|
hsa04919 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|---|---|
| CNPY4 | 6.637 | 6.7676 | 6.4956 | 5.5644 | 5.5122 | 5.4305 | 5.6989 | 5.7247 | 5.6121 |
| PTCD2 | 4.7414 | 4.9418 | 5.0669 | 3.8123 | 3.3417 | 3.4922 | 3.6741 | 3.4882 | 3.4684 |
| PSMD4 | 12.2651 | 12.2941 | 12.0861 | 10.6744 | 11.0524 | 10.9854 | 11.2498 | 11.0404 | 10.957 |
| CEBPA | 6.3381 | 6.4141 | 6.083 | 3.419 | 4.4802 | 4.0412 | 4.3302 | 3.9016 | 3.7224 |
| CDK4 | 11.7465 | 12.0281 | 11.8575 | 8.4493 | 9.6294 | 9.4621 | 8.3643 | 9.3944 | 9.499 |
| TP53 | 10.9055 | 10.762 | 10.5279 | 7.6336 | 7.4155 | 7.2951 | 7.4392 | 7.1925 | 7.9771 |
| MUC1 | 7.6467 | 6.8981 | 7.0264 | 4.2403 | 4.06 | 4.4897 | 4.2713 | 4.34 | 4.3993 |
| AURKB | 10.0324 | 9.792 | 9.903 | 6.439 | 6.996 | 6.2844 | 6.8457 | 7.9253 | 7.4147 |
| CDC25B | 12.0765 | 11.7604 | 12.0577 | 9.2244 | 9.8177 | 9.5007 | 9.9795 | 9.6096 | 9.7486 |
| ICAM3 | 8.225 | 8.8102 | 8.2446 | 6.6038 | 6.0913 | 6.3198 | 6.082 | 5.8808 | 6.1239 |
| BDH1 | 9.2813 | 9.38 | 9.4094 | 5.5855 | 5.9575 | 6.3597 | 6.8907 | 5.9978 | 6.1327 |
| EML3 | 8.988 | 8.6768 | 9.0028 | 7.2903 | 7.2163 | 7.682 | 7.4917 | 7.5085 | 7.4518 |
| ARFIP2 | 9.4745 | 9.424 | 10.3561 | 7.1298 | 6.65 | 6.8119 | 7.9926 | 6.9295 | 6.8266 |
| CDK2 | 9.7594 | 9.787 | 9.8097 | 8.5471 | 8.1903 | 7.9149 | 8.0162 | 8.1082 | 8.1581 |
| SPAG7 | 9.8722 | 9.8828 | 10.3311 | 7.6844 | 7.7119 | 8.0681 | 8.3988 | 8.3797 | 7.7417 |
| MRPL12 | 12.8179 | 12.6801 | 12.9642 | 9.4153 | 9.5186 | 9.254 | 10.3933 | 9.3535 | 10.1674 |
| CBR1 | 8.7159 | 8.9877 | 8.9794 | 7.549 | 7.2812 | 7.55 | 7.5694 | 7.1873 | 7.2396 |
| POP4 | 10.5067 | 10.2807 | 10.4585 | 7.274 | 5.9891 | 7.1669 | 7.7674 | 7.0791 | 7.5849 |
| PSMG1 | 11.1535 | 11.2271 | 11.3566 | 8.6907 | 8.5033 | 9.0259 | 9.2339 | 8.8576 | 9.1637 |
| MYC | 10.0831 | 10.114 | 10.0541 | 8.6128 | 9.1117 | 9.011 | 9.0535 | 8.6735 | 8.8573 |
| CIRBP | 9.2629 | 9.1751 | 8.7853 | 7.0796 | 7.4386 | 7.49 | 7.0206 | 7.4275 | 7.6626 |
| SKIV2L | 8.4716 | 8.3629 | 8.7367 | 5.8165 | 6.1143 | 6.077 | 6.6237 | 6.4424 | 6.4028 |
| STX4 | 9.4398 | 8.9612 | 9.3769 | 7.8403 | 7.4023 | 7.338 | 7.8795 | 7.6956 | 7.772 |
| HADH | 10.2376 | 10.239 | 10.1744 | 8.8799 | 8.7182 | 8.495 | 7.824 | 8.6371 | 8.6102 |
| GALE | 10.1185 | 10.0173 | 9.9016 | 4.8576 | 6.4143 | 5.5954 | 6.4049 | 6.2181 | 5.7831 |
| LOXL1 | 7.8982 | 7.3389 | 7.7443 | 5.6948 | 5.6071 | 5.4198 | 6.1155 | 5.7402 | 6.4028 |
| PAFAH1B3 | 11.0417 | 10.9631 | 11.1535 | 7.8646 | 8.9645 | 8.7379 | 9.1105 | 8.514 | 8.7698 |
| POLD4 | 8.8909 | 8.9561 | 8.8055 | 6.0618 | 6.4143 | 6.7095 | 6.4624 | 6.5037 | 6.8678 |
| TSEN2 | 8.6623 | 8.7199 | 8.514 | 7.0294 | 6.924 | 6.7662 | 6.295 | 6.5785 | 6.3379 |
| FPGS | 8.6184 | 8.4719 | 8.562 | 6.6357 | 6.03 | 6.0349 | 5.936 | 6.234 | 6.6251 |
| PLSCR3 | 8.0119 | 8.1714 | 8.678 | 5.2394 | 6.2662 | 5.8211 | 6.0094 | 5.8633 | 5.7919 |
| COPS7A | 10.1913 | 10.1951 | 10.3156 | 8.3668 | 8.6973 | 8.864 | 8.7448 | 8.574 | 8.7126 |
| CHMP6 | 5.5566 | 5.9406 | 6.105 | 3.8032 | 3.9851 | 3.9275 | 4.0816 | 4.1358 | 3.9002 |
| ST6GALNAC2 | 7.5921 | 8.0114 | 8.7255 | 4.5504 | 5.5355 | 4.894 | 5.4305 | 5.6502 | 5.3053 |
| AMDHD2 | 9.6621 | 9.722 | 10.6744 | 5.9727 | 5.928 | 5.8689 | 6.0965 | 6.1439 | 6.2355 |
| C2CD5 | 10.4144 | 10.2571 | 10.882 | 8.17 | 7.9506 | 8.2827 | 8.7856 | 7.9946 | 7.867 |
| MRPS16 | 11.7838 | 12.0465 | 11.9611 | 10.317 | 10.0633 | 10.3855 | 10.2293 | 9.4538 | 10.213 |
| NOL3 | 9.8144 | 9.8355 | 10.1883 | 8.0483 | 7.8996 | 7.7797 | 8.1017 | 8.3027 | 8.1234 |
| INTS3 | 8.4521 | 8.3823 | 8.7048 | 6.7481 | 6.7569 | 6.9146 | 7.301 | 6.9573 | 6.9586 |
| POLE2 | 8.6447 | 9.5114 | 9.8493 | 6.4997 | 5.519 | 5.7785 | 5.9849 | 5.8745 | 6.4457 |
| CDC45 | 9.4252 | 9.7098 | 9.4252 | 7.217 | 7.0791 | 6.9425 | 6.5855 | 7.252 | 7.458 |
| ECHDC1 | 7.5694 | 7.418 | 7.6747 | 5.914 | 6.1058 | 6.2276 | 6.5059 | 6.0325 | 6.1141 |
| KDELR3 | 6.1126 | 5.7523 | 6.2698 | 3.5323 | 3.5831 | 3.5303 | 4.3669 | 3.5244 | 3.7048 |
| CCT7 | 12.1077 | 11.5304 | 11.7892 | 10.4414 | 10.3636 | 10.1625 | 10.601 | 10.4757 | 10.2495 |
| HAX1 | 10.8643 | 10.3923 | 10.601 | 9.0899 | 9.3895 | 8.9953 | 9.0595 | 8.7864 | 9.181 |
| HNRNPAB | 13.154 | 13.3682 | 13.3244 | 11.1825 | 11.5742 | 11.258 | 11.6177 | 11.2626 | 11.6284 |
| EIF4A3 | 9.5413 | 9.6096 | 9.6375 | 6.8484 | 7.2867 | 7.791 | 7.2818 | 7.411 | 7.3399 |
| RUVBL2 | 11.3237 | 11.8211 | 12.353 | 9.8015 | 10.1297 | 9.7763 | 9.9559 | 10.1476 | 9.9056 |
| SRM | 11.6695 | 11.5366 | 11.3353 | 9.6815 | 10.0472 | 10.0185 | 10.4715 | 9.8857 | 10.1756 |
| PHB2 | 12.5896 | 12.5242 | 12.5001 | 10.8656 | 11.1049 | 10.781 | 11.0453 | 11.0538 | 11.2005 |
| SRSF1 | 8.4981 | 8.1979 | 7.8929 | 6.7274 | 6.3677 | 6.696 | 6.8502 | 6.8876 | 6.8879 |
| PRKAG1 | 8.4406 | 8.574 | 8.4525 | 6.3207 | 7.2038 | 7.0041 | 6.9411 | 6.9325 | 6.6024 |
| IMPDH2 | 9.5066 | 9.243 | 9.6717 | 7.1599 | 7.9408 | 7.4537 | 7.6045 | 7.2045 | 7.5664 |
| FIBP | 10.3043 | 10.4144 | 10.1878 | 8.5515 | 8.9408 | 8.8374 | 9.1206 | 8.7355 | 8.6285 |
| MTHFD1 | 11.6159 | 11.4101 | 12.1733 | 9.6686 | 10.4156 | 9.7046 | 10.1413 | 9.9387 | 9.676 |
| DHFR | 10.5526 | 10.8239 | 10.0717 | 8.9062 | 8.6154 | 8.3151 | 8.0394 | 8.6563 | 8.3364 |
| GDE1 | 8.4734 | 8.5645 | 8.558 | 6.9119 | 7.0666 | 7.0882 | 7.2332 | 7.1866 | 7.5632 |
| CAD | 9.1019 | 9.1808 | 9.0745 | 6.7463 | 7.1667 | 7.5141 | 7.8716 | 7.3754 | 7.4712 |
| RNASEH2A | 11.5706 | 11.5883 | 11.3751 | 7.8435 | 8.1863 | 8.4468 | 7.7744 | 8.4053 | 8.3925 |
| MRPL40 | 9.2321 | 9.1065 | 9.653 | 7.7072 | 7.8059 | 7.0664 | 7.0717 | 6.7775 | 6.8391 |
| TPBG | 11.359 | 11.1689 | 11.8291 | 9.1047 | 9.1309 | 8.4706 | 9.2296 | 9.3146 | 8.5828 |
| PCYOX1 | 7.333 | 7.2196 | 6.8791 | 5.3705 | 5.2501 | 5.8763 | 5.3463 | 5.6344 | 5.4216 |
| NQO2 | 9.3418 | 9.7497 | 9.334 | 6.5739 | 7.9452 | 7.1452 | 7.3131 | 7.0886 | 6.4777 |
| NMI | 6.5729 | 6.1631 | 6.5358 | 3.8803 | 4.5601 | 3.9508 | 3.898 | 4.5492 | 4.4215 |
| GADD45G | 6.3658 | 6.2579 | 5.4822 | 3.9374 | 4.1117 | 4.4057 | 4.1211 | 4.3321 | 4.2243 |
| MRPL57 | 13.2234 | 12.8067 | 13.2586 | 11.1509 | 11.319 | 10.7499 | 10.6381 | 11.0574 | 10.9174 |
| FAM216A | 9.3155 | 8.7771 | 8.6932 | 5.9971 | 5.9578 | 6.2972 | 5.9707 | 5.7398 | 6.1452 |
| NUDT1 | 7.6929 | 7.9107 | 7.9174 | 6.6222 | 6.5588 | 6.4746 | 6.6944 | 6.5872 | 6.4117 |
| POP5 | 12.2577 | 12.9007 | 12.168 | 9.6428 | 10.0594 | 10.277 | 9.9349 | 9.7342 | 9.826 |
| OCEL1 | 7.4769 | 7.4355 | 7.6239 | 6.2375 | 6.5809 | 6.5266 | 6.5101 | 6.4703 | 6.3548 |
| ZDHHC24 | 9.6455 | 9.3623 | 9.4848 | 7.6929 | 7.9118 | 7.6479 | 8.159 | 8.0261 | 8.2004 |
| CCDC22 | 8.8753 | 8.9027 | 8.841 | 6.9336 | 7.4948 | 7.155 | 7.2635 | 7.6076 | 7.5963 |
| GCDH | 8.5415 | 8.1364 | 8.4911 | 6.1723 | 6.2945 | 5.4611 | 6.058 | 6.0701 | 6.2271 |
| UCP2 | 10.7955 | 11.3438 | 11.5366 | 8.5895 | 8.8123 | 8.9376 | 8.812 | 9.3565 | 8.9083 |
| NHP2 | 15 | 14.2958 | 14.8977 | 12.009 | 12.5225 | 12.0513 | 12.5328 | 12.0425 | 12.5001 |
| COMMD4 | 10.0941 | 10.1101 | 10.5725 | 8.0413 | 8.3841 | 8.0966 | 8.3078 | 7.4897 | 8.0623 |
| NDUFAF3 | 8.8693 | 9.7018 | 9.1661 | 7.4693 | 6.893 | 6.9758 | 7.64 | 6.8625 | 6.4984 |
| MPDU1 | 10.4207 | 9.7889 | 9.8551 | 8.3395 | 8.5356 | 7.9665 | 8.0545 | 8.6525 | 8.4739 |
| WDR18 | 9.2741 | 9.4551 | 9.2095 | 7.766 | 7.9852 | 7.3386 | 8.0233 | 7.8618 | 7.6832 |
| PCYT2 | 8.9893 | 8.489 | 8.7298 | 7.1333 | 7.2363 | 6.6368 | 7.3481 | 7.3309 | 7.1747 |
| BUB1 | 9.2838 | 9.0885 | 9.5336 | 7.4861 | 7.8916 | 7.659 | 7.5188 | 8.1274 | 7.9373 |
| CDKN3 | 11.5776 | 11.8874 | 11.2717 | 10.0653 | 9.7098 | 9.9235 | 9.8664 | 9.637 | 9.8385 |
| APEX1 | 14.0835 | 13.2271 | 13.4953 | 11.117 | 11.5441 | 11.0268 | 10.8343 | 11.1313 | 11.7429 |
| SSNA1 | 9.7126 | 9.5802 | 9.6502 | 8.6406 | 8.4154 | 8.3859 | 8.6837 | 8.3711 | 8.2382 |
| GBAP1 | 10.5044 | 10.3176 | 10.3876 | 7.623 | 8.7314 | 8.0945 | 7.7847 | 8.3193 | 7.9837 |
| DOK1 | 6.3871 | 6.3506 | 6.4844 | 4.7913 | 4.9016 | 5.2902 | 5.3566 | 4.764 | 5.0419 |
| GYG1 | 11.5119 | 11.5066 | 11.0315 | 9.5212 | 9.9434 | 9.7311 | 9.4469 | 9.409 | 9.5763 |
| MRPS27 | 9.9585 | 9.8682 | 9.7796 | 8.2311 | 8.7057 | 8.2297 | 8.8096 | 8.3764 | 8.6504 |
| MTG1 | 9.0842 | 9.3376 | 9.4354 | 7.5561 | 7.554 | 7.5947 | 7.2196 | 7.4366 | 7.5029 |
| GADD45GIP1 | 10.6975 | 10.6508 | 10.8719 | 8.9403 | 8.625 | 8.78 | 9.1117 | 8.5995 | 8.7392 |
| PFAS | 9.6081 | 9.5143 | 9.4872 | 6.958 | 7.8657 | 7.774 | 7.5415 | 7.6576 | 7.7506 |
| HSD17B8 | 7.1104 | 7.1809 | 7.0827 | 4.7626 | 4.8382 | 4.466 | 5.3889 | 4.2948 | 4.1311 |
| BMPR1A | 8.5033 | 8.3353 | 8.7054 | 6.2577 | 6.7807 | 7.0772 | 6.0085 | 6.3053 | 6.6292 |
| TUBB4B | 12.1996 | 12.3669 | 12.7782 | 10.0594 | 10.2394 | 10.6765 | 10.0308 | 10.2596 | 9.7022 |
| RBMX | 10.4037 | 10.0141 | 10.0359 | 8.5301 | 8.7825 | 8.0091 | 8.6496 | 8.6042 | 8.7871 |
| SRSF7 | 11.6072 | 11.9936 | 11.338 | 9.472 | 9.2392 | 10.0051 | 9.0115 | 9.4995 | 9.5719 |
| FARSA | 9.3282 | 9.425 | 9.4286 | 7.1402 | 7.878 | 7.1334 | 7.6017 | 7.3972 | 7.9215 |
| MTCH2 | 11.5354 | 11.4999 | 11.5403 | 10.0348 | 9.4534 | 9.0999 | 9.6068 | 8.8841 | 9.1213 |
| GPS1 | 10.9198 | 11.0364 | 11.296 | 9.5189 | 9.3906 | 9.2134 | 9.4918 | 9.4484 | 9.984 |
| MAPKAP1 | 7.7343 | 7.6381 | 7.5594 | 5.5339 | 5.7255 | 6.2785 | 6.3727 | 5.7951 | 5.72 |
| MRPS7 | 12.6247 | 12.4349 | 12.576 | 11.419 | 11.3048 | 11.1652 | 11.1758 | 11.0833 | 11.4439 |
| COA3 | 11.8611 | 11.6193 | 11.9357 | 9.6185 | 10.0132 | 9.6029 | 9.8289 | 10.0103 | 10.1625 |
| COMMD3 | 9.1241 | 9.3358 | 9.5984 | 6.6605 | 7.3836 | 7.5883 | 7.5644 | 6.74 | 7.0906 |
| MRPS34 | 10.967 | 10.8656 | 10.9707 | 9.6521 | 9.9026 | 9.577 | 10.0087 | 9.7872 | 9.5749 |
| ASF1B | 10.9208 | 10.9132 | 10.6122 | 8.6586 | 8.431 | 8.4021 | 8.8176 | 8.6926 | 9.059 |
| C12orf10 | 10.2441 | 10.2992 | 10.2814 | 9.1743 | 9.2281 | 9.1543 | 9.1044 | 9.243 | 9.1272 |
| RMND5B | 9.5138 | 9.396 | 9.747 | 7.7564 | 7.7529 | 7.2846 | 8.05 | 8.1344 | 7.8436 |
| TACC3 | 9.7777 | 9.4378 | 9.365 | 6.7724 | 6.8769 | 7.0901 | 7.2865 | 7.7445 | 7.2646 |
| ZWILCH | 8.7458 | 9.3562 | 9.3581 | 7.2433 | 7.0093 | 6.847 | 7.4053 | 6.9799 | 6.5775 |
| HAUS4 | 9.7703 | 9.3063 | 9.7146 | 6.7219 | 7.2924 | 7.8216 | 7.5689 | 7.5109 | 7.8051 |
| GPN3 | 9.2908 | 9.4373 | 9.5739 | 8.1436 | 7.983 | 7.5899 | 7.7441 | 7.51 | 7.8199 |
| THAP7 | 8.1341 | 7.9413 | 8.2782 | 6.8931 | 7.052 | 6.7561 | 6.9929 | 6.622 | 6.8024 |
| SNRNP25 | 11.9215 | 11.7649 | 12.5791 | 9.7886 | 10.3719 | 9.8724 | 10.2529 | 10.1869 | 10.4321 |
| TMEM134 | 9.183 | 8.9413 | 8.6913 | 7.1257 | 7.059 | 6.7919 | 6.8311 | 6.2326 | 6.8175 |
| ORMDL2 | 11.0758 | 11.3289 | 10.9631 | 9.0035 | 9.5086 | 9.3542 | 8.5966 | 9.0487 | 8.5828 |
| TTC31 | 8.121 | 8.0704 | 8.3776 | 6.437 | 6.5623 | 6.6061 | 6.7768 | 6.2966 | 6.4956 |
| ASPSCR1 | 9.2091 | 8.9399 | 9.0263 | 7.2122 | 7.7076 | 7.1813 | 7.412 | 7.5043 | 7.4136 |
| PBK | 9.5701 | 10.2151 | 9.9141 | 7.7885 | 7.3531 | 7.012 | 7.8292 | 6.8406 | 7.5786 |
| ELP5 | 11.0037 | 10.8459 | 10.8617 | 9.6409 | 8.8085 | 9.0487 | 9.1588 | 8.9013 | 8.927 |
| ATRAID | 10.8743 | 10.9955 | 10.8221 | 9.48 | 9.8769 | 9.8588 | 9.5399 | 9.3434 | 9.4394 |
| ALG6 | 8.6516 | 8.9245 | 8.6102 | 6.0203 | 6.9275 | 6.5925 | 6.5012 | 6.3783 | 6.7864 |
| SH3D21 | 5.9273 | 6.0157 | 5.8172 | 4.3766 | 4.5332 | 4.7803 | 4.5502 | 4.6821 | 4.4285 |
| MANSC1 | 10.5241 | 10.2708 | 10.8729 | 8.5504 | 7.7056 | 7.9094 | 8.3962 | 8.5305 | 8.1004 |
| PUS7L | 6.6011 | 6.4746 | 6.4666 | 5.2094 | 5.4259 | 5.1213 | 5.4108 | 5.1975 | 4.9375 |
| TMCO6 | 7.3681 | 7.1308 | 6.9339 | 4.1795 | 5.1927 | 4.6543 | 5.0965 | 4.6922 | 4.2457 |
| INO80B | 8.2034 | 7.935 | 7.885 | 6.9421 | 6.8854 | 6.4469 | 6.6837 | 6.8148 | 6.5319 |
| CDCA3 | 10.5994 | 10.3149 | 10.0644 | 8.5424 | 8.8568 | 8.3062 | 8.5342 | 8.6649 | 8.7906 |
| CDCA8 | 10.5075 | 10.535 | 10.6957 | 9.7408 | 9.3063 | 9.1138 | 9.3358 | 9.2155 | 9.3172 |
| GINS2 | 10.0902 | 11.0545 | 11.1593 | 7.4585 | 8.3629 | 8.0256 | 8.125 | 8.0854 | 7.7636 |
| PIGP | 9.007 | 9.9692 | 9.716 | 6.5454 | 7.7852 | 6.8416 | 6.6314 | 6.6894 | 6.8856 |
| MCM4 | 10.6872 | 10.8072 | 10.9995 | 9.337 | 9.4311 | 9.2519 | 9.2502 | 8.9799 | 9.5504 |
| FAHD2A | 9.5284 | 9.1535 | 9.3012 | 7.9204 | 7.5485 | 7.8811 | 7.6893 | 7.7191 | 7.6825 |
PAF1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: PAF1
[1] PD2/PAF1 at the Crossroads of the Cancer Network.
[2] PAF1 regulation of promoter-proximal pause release via enhancer activation.
[3] Emerging Insights into the Roles of the Paf1 Complex in Gene Regulation.
[5] Cigarette Smoke Induces Stem Cell Features of Pancreatic Cancer Cells via PAF1.
[7] PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II.
[10] The Paf1 Complex Broadly Impacts the Transcriptome of Saccharomyces cerevisiae.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below