MEN1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN4305
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: daunorubicin
condition dose: 500 nM
condition time: 6 h
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: MEN1
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
TOP2A 
|
hsa01524 
|
|
TOP2B |
hsa01524 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| LIPE | 5.3688 | 5.2385 | 5.6159 | 4.1762 | 4.4259 | 4.3139 |
| PGR | 7.262 | 6.5642 | 7.1251 | 6.0793 | 5.7819 | 5.7285 |
| KLK10 | 3.2531 | 4.2468 | 4.3333 | 3.1005 | 2.1819 | 2.2839 |
| RFX7 | 7.1502 | 6.5796 | 6.3632 | 6.0223 | 5.1501 | 5.3996 |
| MCM3AP-AS1 | 7.0002 | 6.9141 | 6.8763 | 6.0431 | 5.3259 | 5.5346 |
| TICAM1 | 5.3965 | 5.6829 | 5.555 | 4.9102 | 3.3732 | 4.257 |
| MAT2A | 12.4898 | 12.623 | 12.5364 | 11.4028 | 11.1268 | 11.5904 |
| EIF4G1 | 9.8238 | 8.7602 | 8.9897 | 7.8148 | 7.9274 | 8.3396 |
| AURKB | 10.6327 | 10.3221 | 10.5312 | 9.659 | 9.236 | 9.6573 |
| AURKA | 7.5374 | 8.2561 | 8.077 | 6.8682 | 5.2438 | 6.1553 |
| TFDP1 | 9.8165 | 9.642 | 10.0957 | 9.1683 | 8.9735 | 8.9406 |
| CDC20 | 13.8846 | 14.1124 | 13.9658 | 13.1658 | 12.5693 | 12.6936 |
| PLK1 | 10.7957 | 11.3466 | 11.048 | 9.1733 | 7.6371 | 8.1383 |
| IGF1R | 9.0383 | 10.3601 | 9.4873 | 8.2311 | 6.7916 | 7.803 |
| SPTLC2 | 5.0903 | 6.1432 | 5.6104 | 4.8814 | 4.026 | 3.8148 |
| GABPB1 | 9.9316 | 10.2787 | 9.995 | 9.1445 | 8.4187 | 7.5016 |
| BDH1 | 9.5641 | 9.9797 | 9.9637 | 9.1248 | 8.7491 | 8.6312 |
| PDS5A | 9.239 | 9.3871 | 9.7681 | 8.8711 | 7.7594 | 8.3396 |
| XBP1 | 12.4351 | 13.0656 | 13.0912 | 12.0096 | 10.6273 | 11.0721 |
| UBE2C | 12.391 | 12.8582 | 12.8613 | 11.7176 | 10.8645 | 11.2336 |
| DNAJB1 | 12.7884 | 11.4016 | 11.4436 | 10.1959 | 10.0069 | 9.9228 |
| NUP62 | 8.6013 | 9.3719 | 9.1911 | 7.6629 | 6.4451 | 6.9676 |
| SMC4 | 9.3384 | 9.8375 | 9.6503 | 8.6763 | 7.9555 | 8.0926 |
| ARHGEF2 | 6.6786 | 6.2906 | 6.459 | 4.9966 | 3.6384 | 4.7336 |
| CAPN1 | 6.8602 | 6.767 | 6.3932 | 5.7533 | 5.0137 | 5.2458 |
| NOLC1 | 10.5124 | 10.581 | 10.966 | 9.7142 | 8.6997 | 8.8552 |
| RRP1B | 8.8633 | 9.0148 | 9.3098 | 7.993 | 7.4021 | 7.1571 |
| TSEN2 | 9.7285 | 9.8607 | 8.8037 | 8.0373 | 7.1467 | 7.6992 |
| JMJD6 | 11.3099 | 12.0776 | 11.734 | 10.5437 | 10.3786 | 10.192 |
| CLPX | 9.4485 | 9.5681 | 9.84 | 8.8213 | 8.241 | 7.4464 |
| EIF5 | 11.1994 | 11.426 | 11.3973 | 10.6578 | 10.318 | 10.2559 |
| ZDHHC6 | 8.0586 | 7.8356 | 8.1812 | 7.3779 | 6.5253 | 5.9273 |
| INTS3 | 8.3995 | 8.7096 | 8.4646 | 7.2982 | 6.4909 | 7.2271 |
| CYB561 | 10.2057 | 10.1106 | 10.7177 | 8.708 | 9.4675 | 9.3983 |
| GATA3 | 10.2167 | 10.8625 | 11.0056 | 9.3713 | 8.5166 | 9.6196 |
| PARP2 | 9.3964 | 10.087 | 9.9565 | 8.5389 | 5.2255 | 5.9802 |
| BAG3 | 12.9887 | 12.931 | 13.1494 | 12.375 | 12.247 | 11.948 |
| EVL | 7.2071 | 8.1505 | 8.1892 | 6.6549 | 6.0251 | 5.6364 |
| POLE2 | 9.1723 | 9.413 | 9.495 | 8.4516 | 8.3873 | 7.4567 |
| DLGAP5 | 10.2125 | 10.3649 | 10.5947 | 8.8175 | 7.5892 | 8.709 |
| ZNF146 | 10.576 | 10.672 | 11.2332 | 10.228 | 9.4235 | 9.0198 |
| NOP56 | 13.4626 | 13.6651 | 13.8846 | 12.4145 | 11.7874 | 12.9259 |
| IPO7 | 10.7122 | 11.1048 | 10.6158 | 9.9316 | 9.2517 | 9.6845 |
| HNRNPF | 11.2575 | 11.4289 | 11.0706 | 10.5459 | 9.8803 | 10.0719 |
| PSMA3 | 13.4078 | 13.2535 | 13.1938 | 12.3239 | 11.8869 | 12.4023 |
| PSMC6 | 13.7533 | 13.5947 | 13.8014 | 12.0082 | 11.6229 | 11.806 |
| ADNP | 9.3792 | 9.3699 | 9.8361 | 8.3426 | 8.9305 | 7.7665 |
| SLC39A6 | 13.0402 | 13.1154 | 14.5306 | 12.3102 | 9.7351 | 10.3508 |
| TACSTD2 | 10.9421 | 11.318 | 11.7126 | 9.5656 | 8.2484 | 9.0206 |
| SNRPB2 | 12.0965 | 12.0566 | 12.2768 | 11.0894 | 11.0875 | 11.1163 |
| SNRPD1 | 12.9259 | 12.9055 | 12.8402 | 11.5426 | 11.0744 | 11.159 |
| PLCG1 | 9.9412 | 10.0168 | 9.4004 | 8.7565 | 8.3657 | 8.4252 |
| SS18 | 8.6061 | 8.4746 | 8.9715 | 8.1708 | 7.2263 | 7.0071 |
| MTIF2 | 10.1572 | 9.6245 | 9.7411 | 9.1202 | 8.3316 | 8.6328 |
| SPAG5 | 8.469 | 8.7643 | 8.709 | 7.6936 | 6.768 | 7.4583 |
| NOTCH3 | 6.2445 | 6.2864 | 7.0108 | 5.3065 | 4.1637 | 5.2987 |
| PPRC1 | 9.7873 | 9.0749 | 9.2566 | 8.3819 | 7.6725 | 8.0325 |
| VRK1 | 9.4226 | 10.086 | 9.6384 | 8.6637 | 9.0328 | 8.2022 |
| NLE1 | 9.0799 | 8.4804 | 8.8289 | 7.8682 | 7.757 | 7.9008 |
| RFC4 | 10.4341 | 10.9242 | 11.2729 | 9.8515 | 9.7745 | 10.0473 |
| THOC1 | 9.4579 | 9.7326 | 9.3449 | 8.5161 | 8.2596 | 8.6024 |
| GTSE1 | 9.4221 | 9.239 | 9.3249 | 8.1035 | 7.6671 | 8.278 |
| KIF11 | 10.0918 | 9.7845 | 9.639 | 8.9699 | 9.006 | 8.8076 |
| KIF23 | 10.0502 | 8.7703 | 9.7395 | 8.2533 | 7.7274 | 6.6875 |
| WDHD1 | 7.324 | 6.7645 | 7.0929 | 6.014 | 5.6301 | 5.7516 |
| ALDH3B2 | 7.3166 | 8.2236 | 7.1916 | 6.2383 | 6.1576 | 6.4238 |
| TFF1 | 14.4696 | 14.8332 | 15 | 13.1288 | 9.553 | 12.218 |
| NPM3 | 11.1712 | 11.2674 | 11.3847 | 10.1234 | 9.9482 | 10.626 |
| KIF20B | 7.6103 | 7.9137 | 8.0992 | 7.1156 | 6.6515 | 7.107 |
| PFDN4 | 14.2235 | 13.5771 | 14.0246 | 13.4114 | 12.0544 | 12.0965 |
| PDZK1 | 6.1414 | 6.4542 | 7.8673 | 5.6504 | 3.8985 | 4.5778 |
| ATXN3 | 8.3011 | 7.8472 | 8.0197 | 7.3591 | 6.5372 | 6.5558 |
| KIN | 9.6971 | 9.0524 | 9.4982 | 8.2438 | 8.1901 | 7.8304 |
| BLM | 5.6347 | 6.0804 | 6.4155 | 5.0583 | 3.9786 | 4.7228 |
| GREB1 | 10.1525 | 9.2435 | 10.7552 | 9.1256 | 7.4171 | 7.5584 |
| HOXC6 | 7.2036 | 6.5 | 6.2241 | 5.1975 | 4.4268 | 5.0134 |
| HSPH1 | 14.4213 | 14.0874 | 14.4696 | 13.4424 | 13.3021 | 13.4594 |
| RBMS1 | 8.8879 | 8.7741 | 8.9166 | 8.0856 | 7.8805 | 7.3225 |
| EXOSC10 | 10.0948 | 9.6269 | 9.7762 | 9.098 | 8.2502 | 8.6902 |
| RPL36AL | 13.4892 | 13.3165 | 13.2653 | 12.3768 | 12.2565 | 12.3578 |
| CENPF | 7.6186 | 7.5417 | 7.727 | 7.0423 | 6.1587 | 6.2318 |
| XPO1 | 10.7293 | 10.9469 | 11.1284 | 10.3967 | 9.7512 | 9.24 |
| SRP72 | 7.7439 | 8.3107 | 8.5198 | 7.2304 | 6.9821 | 6.4592 |
| NR2F2 | 10.6241 | 10.5772 | 11.8206 | 9.7554 | 8.02 | 9.5229 |
| PATZ1 | 8.9952 | 9.0064 | 9.1405 | 7.9688 | 7.6502 | 7.9112 |
| CACNB3 | 9.9575 | 9.5874 | 9.9362 | 9.0275 | 8.7449 | 8.9942 |
| BUB1 | 10.2777 | 9.7414 | 10.3584 | 8.6572 | 7.7915 | 7.8738 |
| UCK2 | 11.6034 | 11.5094 | 11.8393 | 10.3139 | 10.9878 | 10.4321 |
| SPC25 | 9.4803 | 8.2836 | 8.9099 | 7.5585 | 7.4392 | 7.3302 |
| APEX1 | 14.5131 | 13.7391 | 15 | 13.4219 | 13.1189 | 12.2271 |
| TPX2 | 10.01 | 10.6595 | 10.7441 | 9.5868 | 8.7153 | 8.2961 |
| IRX5 | 5.1902 | 5.9845 | 6.1322 | 4.5625 | 3.539 | 3.0589 |
| CACYBP | 13.0338 | 12.785 | 13.4357 | 12.3475 | 12.0408 | 11.3113 |
| DYNC1H1 | 10.7467 | 10.0147 | 11.0769 | 9.7996 | 9.1802 | 9.007 |
| HSP90AA1 | 15 | 14.2443 | 14.3211 | 12.9174 | 12.4816 | 13.2064 |
| SMARCE1 | 10.7492 | 10.9967 | 10.7987 | 10.3396 | 9.1723 | 9.4945 |
| MKI67 | 9.2716 | 8.1939 | 8.3183 | 7.6591 | 6.7195 | 6.9546 |
| PNN | 12.9133 | 13.1744 | 13.2738 | 11.8174 | 12.263 | 10.7278 |
| XPOT | 11.9132 | 12.247 | 12.4436 | 11.5368 | 10.3445 | 10.4767 |
| NRP1 | 8.5098 | 7.8431 | 7.6565 | 6.8634 | 5.9439 | 6.2631 |
| YLPM1 | 9.0821 | 8.6107 | 9.7069 | 8.3153 | 7.1927 | 7.3912 |
| NCAPH | 10.288 | 9.8201 | 9.9024 | 9.069 | 8.5887 | 9.1117 |
| SP3 | 9.1805 | 8.8267 | 9.2945 | 8.1131 | 8.245 | 7.6271 |
| PFAS | 10.0831 | 9.9697 | 9.9174 | 9.4995 | 8.933 | 8.8155 |
| UMPS | 10.3622 | 10.5798 | 10.6322 | 9.5173 | 9.7334 | 9.4568 |
| FAM136A | 10.9508 | 11.0665 | 11.2469 | 10.4063 | 10.0513 | 10.1129 |
| DAAM1 | 9.402 | 9.3108 | 8.666 | 7.8365 | 7.4227 | 7.9572 |
| SKA1 | 7.8206 | 7.2358 | 7.5205 | 6.8147 | 6.4895 | 6.4869 |
| MRPL16 | 12.1127 | 11.3307 | 11.9697 | 10.9699 | 10.5445 | 10.8366 |
| NMD3 | 10.3967 | 10.7467 | 11.1985 | 9.29 | 8.879 | 8.569 |
| NDC1 | 9.8301 | 9.2416 | 10.1746 | 8.5832 | 8.8309 | 7.4229 |
| GTPBP4 | 12.3435 | 12.4167 | 12.9951 | 11.9719 | 10.7629 | 11.2554 |
| LGR4 | 7.8991 | 7.5908 | 8.4065 | 7.0963 | 6.6901 | 6.8354 |
| ZWILCH | 9.6185 | 10.0737 | 9.9872 | 8.922 | 7.5639 | 7.6032 |
| FNDC3B | 7.065 | 6.9316 | 7.0984 | 6.212 | 5.665 | 4.3693 |
| NCAPG | 9.6004 | 9.4648 | 9.6073 | 8.8223 | 8.2491 | 8.6696 |
| UTP6 | 9.3836 | 8.6773 | 8.9937 | 8.1998 | 6.9664 | 7.8601 |
| WARS2 | 8.7855 | 8.2109 | 8.4445 | 7.431 | 7.6643 | 7.575 |
| CEP192 | 7.2473 | 7.1173 | 7.2336 | 6.4879 | 5.6787 | 6.0157 |
| GMIP | 6.8781 | 6.7627 | 7.1789 | 6.2689 | 5.7744 | 6.1154 |
| RRP15 | 12.0028 | 12.2978 | 12.7298 | 11.3783 | 11.049 | 10.8333 |
| TMEM185B | 8.165 | 7.99 | 8.1266 | 7.3154 | 6.5491 | 6.3246 |
| TIPIN | 8.929 | 9.1476 | 9.6107 | 8.4397 | 6.6665 | 7.4888 |
| PGS1 | 8.9945 | 8.5261 | 8.6657 | 8.118 | 7.2735 | 7.683 |
| TASP1 | 7.9947 | 7.8942 | 8.4184 | 7.4022 | 6.3991 | 6.707 |
| LRFN4 | 7.6143 | 8.0038 | 7.8756 | 7.2074 | 6.7783 | 6.7236 |
| RSRC1 | 9.2605 | 9.2567 | 9.3536 | 8.1215 | 8.0301 | 8.4767 |
| RASIP1 | 6.0558 | 5.1258 | 5.8089 | 4.528 | 3.9202 | 4.2154 |
| PARPBP | 8.0051 | 7.4053 | 7.3902 | 6.5127 | 6.0135 | 6.0496 |
| MCM10 | 10.1091 | 9.2073 | 10.2213 | 8.5301 | 7.6219 | 8.0493 |
| CDCA3 | 10.5778 | 9.6725 | 10.3501 | 9.2799 | 9.1154 | 8.928 |
| BRIP1 | 9.1906 | 9.139 | 8.9932 | 8.4534 | 7.3608 | 7.9468 |
| NOL11 | 13.9423 | 13.4509 | 14.1981 | 13.0611 | 11.5965 | 12.5742 |
| TBC1D16 | 8.0872 | 7.9172 | 8.1714 | 7.3345 | 7.3544 | 6.8972 |
| CENPN | 9.1783 | 8.8302 | 8.9499 | 7.6834 | 7.1257 | 7.5896 |
| DCLRE1C | 6.199 | 6.4743 | 6.7192 | 5.226 | 4.1525 | 4.4978 |
MEN1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: MEN1
[1] Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1).
[2] Multiple endocrine neoplasia type 1 (MEN1) and type 4 (MEN4).
[4] Phenotypes Associated With MEN1 Syndrome: A Focus on Genotype-Phenotype Correlations.
[5] Current and emerging therapies for PNETs in patients with or without MEN1.
[6] The future: genetics advances in MEN1 therapeutic approaches and management strategies.
[7] From Initial Description by Wermer to Present-Day MEN1: What have We Learned?
[8] Recent Topics Around Multiple Endocrine Neoplasia Type 1.
[9] Update on phenotypes of MEN1 mutation carriers.
[10] True MEN1 or phenocopy? Evidence for geno-phenotypic correlations in MEN1 syndrome.
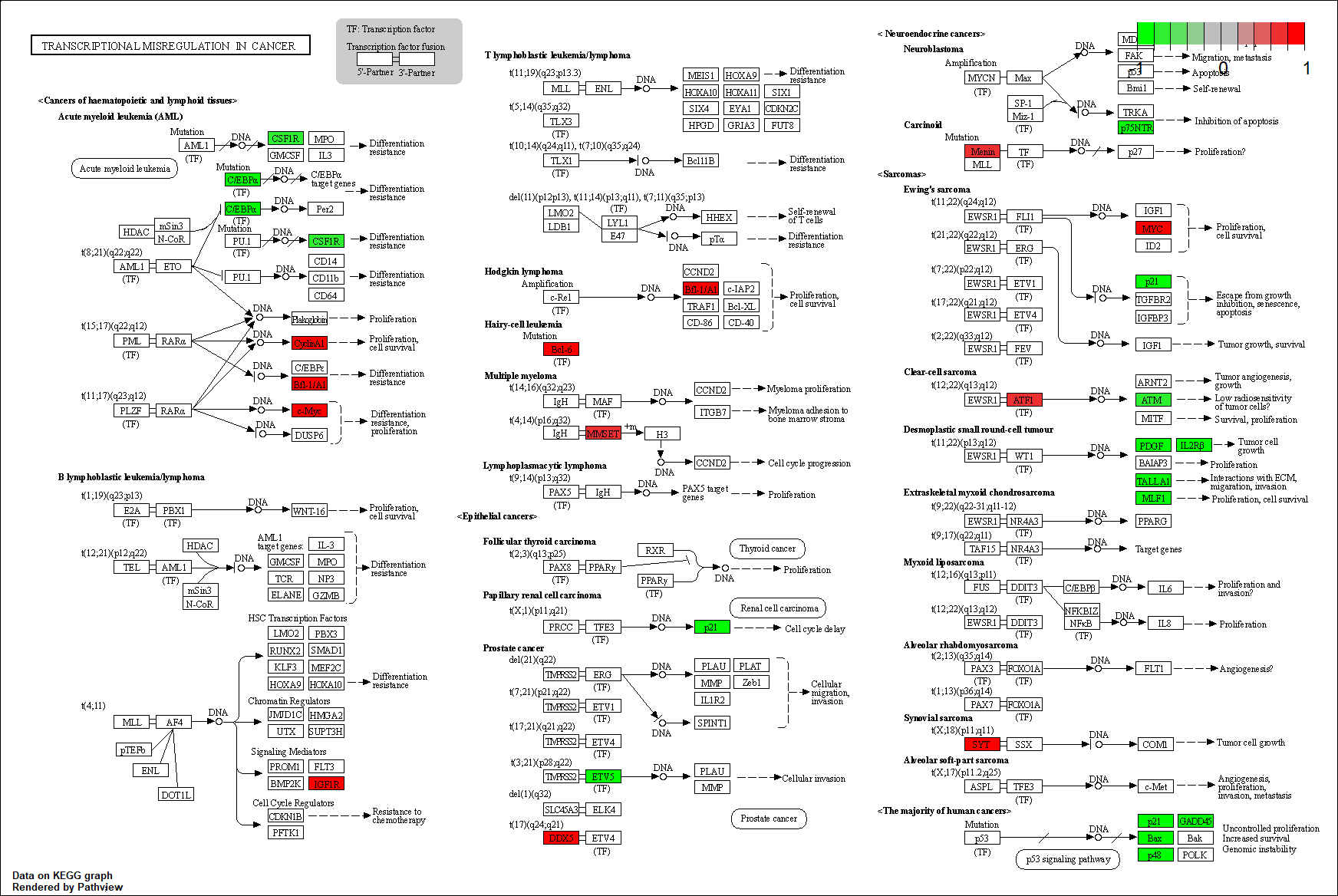
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below
hsa05202