KLF10 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN6735
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: palbociclib
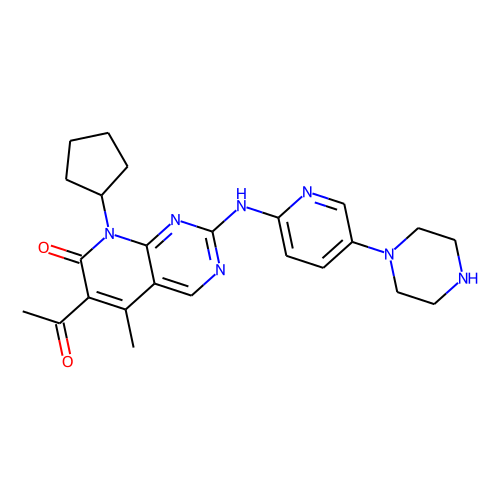
| ID | BRD-K51313569 |
| name | palbociclib |
| type | Small molecule |
| molecular formula | C24H29N7O2 |
| molecular weight | 447.5 |
| condition dose | 3.33 um |
| condition time | 24 h |
| smiles | CC(=O)c1c(C)c2cnc(Nc3ccc(cn3)N3CCNCC3)nc2n(C2CCCC2)c1=O |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: KLF10
Transcription regulatory factors corresponding to this network
ARTICLE: palbociclib
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
CDK4 |
hsa04934 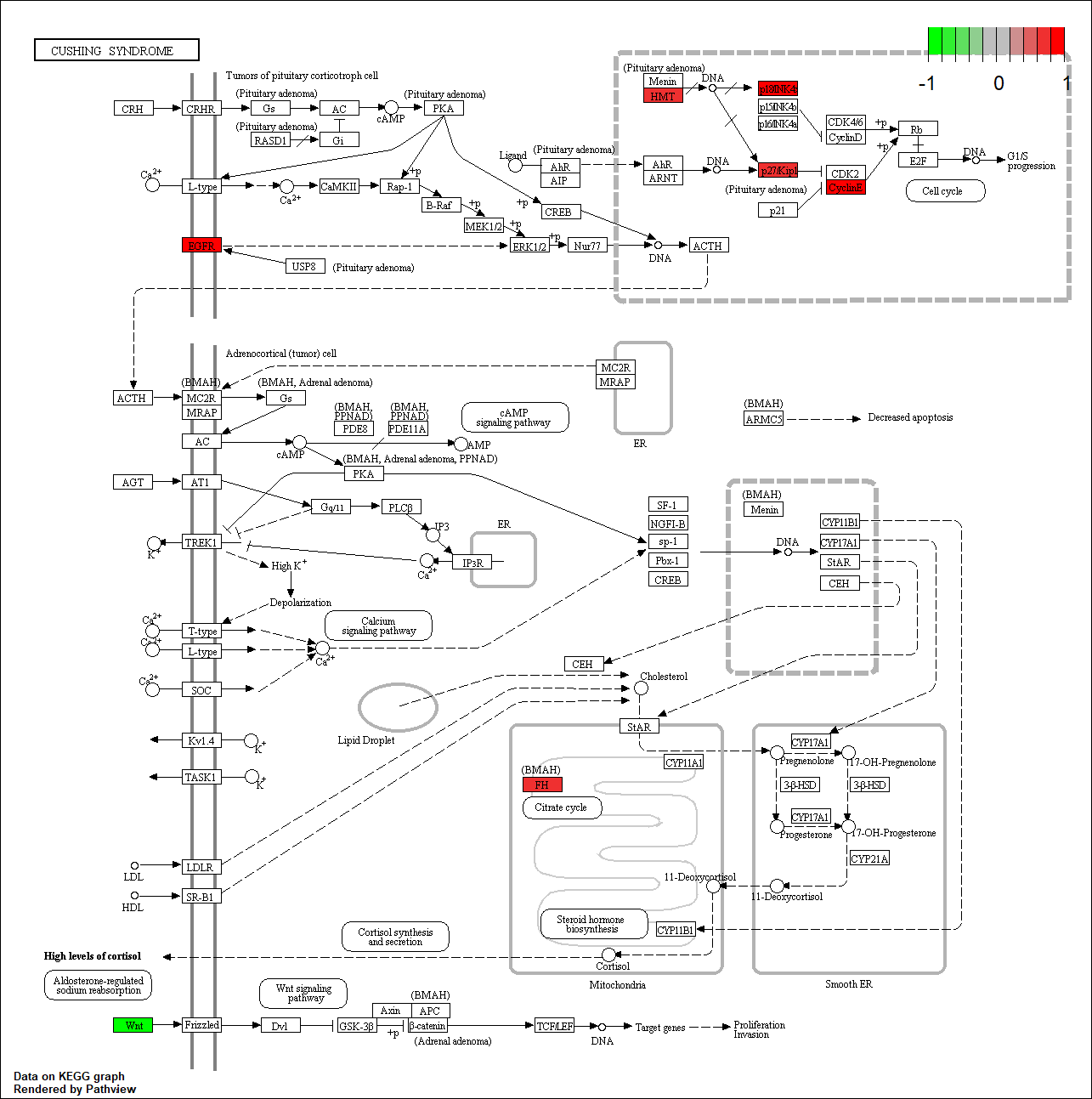
|
|
CDK6 
|
hsa05206 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|
| TACC3 | 8.7889 | 9.3105 | 9.379 | 7.0365 | 7.2481 | 7.2511 | 7.0875 |
| ZWILCH | 9.2768 | 8.6617 | 9.4468 | 8.7683 | 7.3617 | 7.7794 | 7.4782 |
| CARHSP1 | 8.8314 | 8.7617 | 8.5735 | 7.7934 | 7.9544 | 7.4132 | 8.1552 |
| CDCA4 | 7.951 | 7.5775 | 8.0391 | 6.7301 | 6.7697 | 5.8999 | 6.9024 |
| CMC2 | 11.3109 | 11.2885 | 12.729 | 10.6199 | 11.1816 | 9.6569 | 9.6712 |
| JADE1 | 6.0209 | 5.0818 | 5.4867 | 4.6506 | 4.5114 | 4.7162 | 4.4006 |
| SUV39H1 | 6.9762 | 6.8203 | 7.5113 | 5.1194 | 5.7843 | 5.3748 | 6.3 |
| CRTC3 | 5.5352 | 5.8508 | 6.5362 | 5.2836 | 5.0091 | 4.5442 | 4.664 |
| NCAPG | 8.9786 | 8.9381 | 8.7661 | 6.8962 | 7.0595 | 6.9957 | 8.2826 |
| KIF20A | 9.1825 | 9.6288 | 9.746 | 6.4718 | 5.4953 | 3.7716 | 6.6839 |
| NUP107 | 12.0343 | 11.6464 | 11.9849 | 11.4571 | 10.6375 | 10.2948 | 10.7899 |
| ATAD2 | 10.788 | 11.0781 | 10.9969 | 7.9081 | 7.8244 | 7.2987 | 10.4537 |
| SOBP | 6.7822 | 7.0689 | 7.1405 | 6.3877 | 5.9194 | 6.0062 | 6.0573 |
| RMI1 | 8.5269 | 7.9076 | 7.4258 | 7.0319 | 5.94 | 6.8228 | 7.3143 |
| PXMP2 | 10.4504 | 10.3315 | 10.5083 | 9.4113 | 9.7123 | 9.1886 | 8.4665 |
| PBK | 8.6624 | 8.2818 | 9.6141 | 6.6551 | 5.8764 | 7.1789 | 7.9243 |
| TIPIN | 8.7845 | 8.0784 | 7.9776 | 7.4746 | 7.3165 | 7.2281 | 6.8873 |
| KIF15 | 6.9226 | 6.5251 | 6.8505 | 4.4398 | 4.588 | 4.2813 | 6.6304 |
| DSN1 | 7.5365 | 7.5191 | 7.1935 | 6.3085 | 5.7717 | 5.769 | 7.1099 |
| IGFLR1 | 7.3269 | 7.1939 | 7.678 | 6.3847 | 6.2454 | 6.6116 | 6.8833 |
| TDP1 | 7.0112 | 7.1055 | 8.133 | 6.4581 | 6.5353 | 6.0604 | 6.3972 |
| ASPM | 8.7529 | 8.5911 | 8.6114 | 6.8638 | 5.5939 | 6.0954 | 8.4603 |
| MCM10 | 8.7315 | 8.8877 | 8.5165 | 6.8189 | 6.3001 | 6.7452 | 8.7677 |
| ANP32E | 8.5405 | 7.5087 | 7.7918 | 6.9138 | 5.3265 | 6.0547 | 6.1675 |
| CDCA8 | 9.7596 | 10.0904 | 10.1479 | 8.6494 | 8.4424 | 7.8078 | 9.6012 |
| GINS2 | 11.2518 | 10.2745 | 10.4426 | 8.0485 | 8.0692 | 8.0533 | 9.8481 |
| DONSON | 8.2954 | 8.1692 | 7.9437 | 6.6174 | 6.172 | 6.4847 | 7.3798 |
| BRIP1 | 9.5056 | 9.7403 | 9.1543 | 8.4621 | 8.1761 | 7.9979 | 8.2755 |
| GLRX5 | 9.6272 | 9.3972 | 9.9776 | 8.0709 | 8.8734 | 8.9081 | 8.7183 |
| TRMT5 | 14.12 | 13.8872 | 13.1245 | 12.3302 | 11.8049 | 11.8105 | 12.7132 |
| KIF18B | 8.0076 | 8.0336 | 7.6029 | 5.8207 | 5.8688 | 5.0837 | 7.4421 |
| RACGAP1 | 9.9468 | 9.5096 | 10.3045 | 8.7064 | 7.6475 | 7.4433 | 8.0372 |
| CENPN | 8.2743 | 8.429 | 8.2884 | 6.9612 | 6.9515 | 7.0365 | 7.3515 |
| SAFB2 | 8.7043 | 8.4696 | 8.6578 | 7.78 | 7.5674 | 7.4667 | 7.9744 |
| CELSR2 | 10.3416 | 10.4517 | 10.0704 | 9.1178 | 9.707 | 8.2681 | 9.5325 |
| EIF3F | 10.1194 | 10.0367 | 10.4391 | 9.8083 | 8.8506 | 8.6048 | 8.745 |
| PTP4A1 | 11.7125 | 10.3192 | 11.1033 | 10.0175 | 9.3 | 9.8845 | 8.1789 |
| STMN1 | 9.1353 | 8.6863 | 9.2382 | 6.1075 | 6.2607 | 5.7242 | 7.212 |
| PSMD8 | 11.8794 | 12.1928 | 12.6524 | 11.1367 | 11.5196 | 11.4529 | 10.4381 |
| FUS | 9.8521 | 10.5793 | 10.7914 | 9.5757 | 9.5134 | 9.2004 | 8.6818 |
| SRRM1 | 10.8774 | 11.0312 | 11.0301 | 10.353 | 9.9264 | 8.682 | 10.0444 |
| RAF1 | 8.6378 | 9.5509 | 9.6855 | 7.6563 | 8.1268 | 8.5808 | 7.768 |
| TOP2A | 12.0243 | 11.9618 | 11.9679 | 8.5568 | 9.4971 | 8.8751 | 10.6025 |
| ANP32B | 11.9784 | 11.9312 | 11.448 | 10.8507 | 11.4027 | 9.1786 | 9.1506 |
| RRM1 | 12.7301 | 12.0302 | 11.3817 | 9.9063 | 9.6126 | 9.8734 | 11.1886 |
| MCM3 | 11.1995 | 11.3304 | 11.2264 | 10.4275 | 10.1441 | 9.0044 | 10.3754 |
| ICMT | 9.1693 | 9.2677 | 9.365 | 8.4855 | 8.7171 | 7.8513 | 7.8178 |
| BST2 | 5.7052 | 6.0794 | 8.1124 | 4.7032 | 4.4125 | 5.4453 | 3.3353 |
| SMC4 | 10.33 | 10.2656 | 10.4066 | 8.9552 | 8.8171 | 7.9237 | 8.0265 |
| MYBL2 | 11.8834 | 9.8245 | 10.4021 | 6.9619 | 7.6387 | 7.9135 | 10.4753 |
| TUBG1 | 10.589 | 11.4022 | 11.1634 | 9.1514 | 9.6914 | 8.5153 | 10.6243 |
| CDC25B | 11.6367 | 12.3561 | 12.2955 | 10.7421 | 11.4451 | 10.6223 | 10.571 |
| BIRC5 | 10.3086 | 10.666 | 10.7849 | 7.7439 | 7.6913 | 8.2021 | 9.3834 |
| IFRD1 | 12.2296 | 12.7595 | 11.7668 | 11.3536 | 10.8611 | 11.541 | 11.1521 |
| NUP62 | 9.0983 | 9.8186 | 9.4104 | 8.2141 | 8.6889 | 8.1791 | 6.0816 |
| PLK1 | 7.8078 | 8.89 | 9.385 | 6.2726 | 7.4661 | 6.7346 | 7.734 |
| TACSTD2 | 13.5157 | 14.1886 | 13.0397 | 12.7616 | 12.0784 | 10.7712 | 12.5989 |
| UNG | 10.6886 | 11.3983 | 10.465 | 9.6292 | 10.0445 | 9.7525 | 9.4001 |
| USP1 | 6.8486 | 6.712 | 7.0387 | 5.4484 | 5.4138 | 4.7932 | 6.1209 |
| FOXM1 | 11.9103 | 12.2046 | 12.1949 | 9.6688 | 9.2609 | 8.694 | 11.3244 |
| TYMS | 13.6898 | 14.2596 | 11.8064 | 10.0655 | 11.477 | 10.3545 | 10.6057 |
| LIG1 | 8.9462 | 9.8395 | 10.0516 | 6.6784 | 7.5672 | 7.2373 | 6.9483 |
| UBE2S | 10.443 | 12.5015 | 12.4999 | 9.2022 | 10.2215 | 10.4354 | 9.1886 |
| HPRT1 | 11.3139 | 10.7912 | 10.9665 | 10.2294 | 10.0801 | 10.1154 | 9.2303 |
| CDC20 | 11.7546 | 12.6606 | 12.5013 | 9.4398 | 10.0396 | 10.0155 | 10.2293 |
| UBE2C | 11.0273 | 10.9186 | 11.3032 | 7.8222 | 7.8474 | 7.8026 | 8.7479 |
| RNASEH2A | 9.8166 | 9.5813 | 11.1312 | 8.0753 | 7.9845 | 8.0593 | 9.5537 |
| TIMELESS | 10.8811 | 10.4506 | 10.6891 | 9.4354 | 8.9899 | 9.175 | 10.328 |
| MDC1 | 7.5418 | 6.7603 | 7.0492 | 6.534 | 6.14 | 5.3975 | 6.1529 |
| SPAG5 | 7.5283 | 8.2087 | 8.5924 | 6.2451 | 6.8209 | 6.5045 | 7.3763 |
| EZH2 | 8.875 | 9.3002 | 8.4905 | 8.0302 | 7.7231 | 7.2909 | 7.133 |
| MAD2L1 | 9.93 | 10.2341 | 9.6295 | 8.4398 | 7.8633 | 8.1864 | 9.4892 |
| DDB2 | 9.1439 | 9.4029 | 9.9373 | 8.4338 | 8.6561 | 7.878 | 8.6367 |
| CCNA2 | 9.5484 | 8.9849 | 9.2821 | 5.7507 | 5.9885 | 5.7794 | 6.8803 |
| TMPO | 11.2017 | 10.7208 | 10.7009 | 9.3082 | 9.0297 | 7.6885 | 9.4825 |
| SEC61G | 11.7927 | 11.5284 | 12.288 | 11.1378 | 10.9117 | 10.5451 | 9.5161 |
| PTTG1 | 9.8251 | 10.1116 | 10.8444 | 8.446 | 8.2686 | 6.2561 | 8.8662 |
| GGH | 8.2272 | 7.4134 | 7.25 | 6.3979 | 6.0121 | 3.9649 | 6.8528 |
| RFC2 | 8.7075 | 9.3306 | 9.6142 | 7.0316 | 6.888 | 7.2245 | 7.5429 |
| DLGAP5 | 9.3291 | 9.6028 | 9.8323 | 6.7985 | 7.6357 | 7.2176 | 7.9794 |
| VRK1 | 8.332 | 7.8444 | 8.243 | 7.0786 | 7.2024 | 6.5866 | 7.3091 |
| RFC4 | 9.9799 | 9.0542 | 9.5398 | 8.1634 | 7.9528 | 8.3184 | 9.1149 |
| TRIP13 | 10.7014 | 10.8999 | 11.0262 | 8.9541 | 10.0037 | 9.0685 | 9.8769 |
| AURKA | 7.7149 | 7.6574 | 8.2388 | 6.8017 | 6.5655 | 7.1129 | 5.7529 |
| RFC3 | 11.1712 | 10.7242 | 10.6157 | 9.2867 | 8.6532 | 7.8626 | 10.6255 |
| CDKN2C | 7.4223 | 7.3953 | 9.2115 | 5.1779 | 7.2112 | 6.5615 | 6.2826 |
| RBM14 | 8.618 | 9.2235 | 9.6279 | 8.2732 | 8.2353 | 8.4641 | 7.5576 |
| DBF4 | 9.1985 | 8.7362 | 9.2521 | 8.2779 | 7.3801 | 7.3959 | 7.5238 |
| IFI6 | 9.287 | 7.9396 | 8.8218 | 7.8638 | 7.7809 | 6.7065 | 7.6322 |
| BRCA1 | 5.8514 | 6.0607 | 5.3748 | 3.8923 | 3.4091 | 2.9294 | 5.8216 |
| CDC25A | 6.9997 | 7.1909 | 7.4943 | 4.5405 | 4.9629 | 4.6717 | 6.048 |
| NUDT1 | 7.4841 | 8.0847 | 7.8088 | 6.8555 | 6.1464 | 5.8526 | 7.3922 |
| FEN1 | 11.1288 | 11.7022 | 10.6764 | 9.5855 | 8.898 | 9.4859 | 11.0472 |
| CHAF1B | 7.7723 | 7.467 | 7.574 | 6.456 | 6.1862 | 6.3905 | 7.041 |
| PRIM1 | 8.891 | 8.5053 | 8.7763 | 7.6004 | 7.0993 | 6.4498 | 8.0842 |
| TFAP2C | 12.7711 | 13.1082 | 12.1045 | 10.7234 | 11.3084 | 11.2881 | 12.3879 |
| STIL | 7.2479 | 7.1719 | 7.2173 | 5.7897 | 5.3897 | 6.0855 | 6.8821 |
| KLK11 | 9.0231 | 8.4313 | 8.567 | 5.1368 | 5.1117 | 5.2572 | 4.6006 |
| SLC25A40 | 6.7788 | 6.657 | 6.5228 | 6.2324 | 5.7651 | 5.553 | 5.4804 |
| BLM | 5.8717 | 5.3217 | 5.8317 | 4.6131 | 4.0277 | 4.6776 | 4.6685 |
| GREB1 | 12.1919 | 11.554 | 11.153 | 10.9025 | 10.2348 | 10.0576 | 9.0454 |
| POLE2 | 7.6105 | 7.0648 | 7.3994 | 4.0016 | 4.9077 | 4.0546 | 6.8913 |
| SLBP | 12.017 | 12.0027 | 10.6472 | 9.7148 | 10.2689 | 10.242 | 10.7869 |
| GABPB1 | 10.3565 | 9.8898 | 10.2359 | 9.275 | 9.4861 | 9.0421 | 8.101 |
| KIF14 | 6.0659 | 5.4562 | 6.9754 | 4.1838 | 4.0739 | 3.4556 | 3.3216 |
| HMMR | 9.4866 | 9.5162 | 9.7165 | 7.4998 | 8.1671 | 7.057 | 9.5088 |
| BAZ1B | 8.7583 | 8.4405 | 8.7249 | 8.0336 | 7.7241 | 6.9238 | 7.2919 |
| HMGN2 | 13.2096 | 13.2489 | 12.9503 | 12.4141 | 11.7443 | 11.1736 | 12.0429 |
| CLU | 8.0715 | 7.2542 | 9.6817 | 5.4653 | 5.2086 | 7.0869 | 4.482 |
| TUBB | 10.6634 | 11.0987 | 11.159 | 8.9648 | 9.7322 | 10.1034 | 8.1572 |
| PSIP1 | 8.9348 | 8.9958 | 9.0044 | 7.8891 | 7.4864 | 4.3725 | 7.476 |
| CHI3L1 | 10.5393 | 8.4253 | 7.5993 | 6.5832 | 4.324 | 7.5962 | 6.7999 |
| RPA3 | 9.6292 | 8.6044 | 9.2724 | 8.4726 | 6.888 | 8.099 | 7.2306 |
| NCBP1 | 8.6622 | 8.7797 | 8.0421 | 7.5979 | 7.5944 | 7.1787 | 7.7086 |
| EED | 9.698 | 8.5948 | 8.8958 | 8.3516 | 7.7708 | 7.1129 | 7.1859 |
| EYA2 | 4.2607 | 3.8175 | 5.3244 | 3.3607 | 3.431 | 2.3666 | 3.4853 |
| CDKN3 | 9.6796 | 10.1446 | 10.7704 | 8.2749 | 8.1841 | 7.4772 | 7.5154 |
| TPX2 | 9.0051 | 9.9935 | 10.4029 | 7.7325 | 8.1577 | 7.3824 | 8.891 |
| CTSV | 7.9898 | 7.553 | 7.7624 | 5.5956 | 6.365 | 5.7323 | 6.5152 |
| CHEK2 | 7.4276 | 7.8805 | 7.9135 | 6.1511 | 7.0037 | 5.8861 | 6.7929 |
| CACYBP | 11.6908 | 12.2164 | 11.653 | 11.1486 | 10.8563 | 11.0779 | 10.7794 |
| POLR2L | 12.4314 | 12.9071 | 12.5663 | 12.0229 | 11.3218 | 11.1767 | 11.7512 |
| ARL6IP1 | 9.3528 | 9.7533 | 10.2369 | 8.7862 | 7.9898 | 7.0929 | 6.5136 |
| TUBA4A | 10.8697 | 11.1491 | 10.6642 | 9.0614 | 9.2896 | 10.0886 | 10.2614 |
| TMEM97 | 10.7527 | 10.6704 | 10.8993 | 9.7931 | 10.0059 | 9.062 | 7.7248 |
| NUP188 | 8.1965 | 8.5076 | 8.5734 | 7.6026 | 7.6579 | 7.6334 | 7.5759 |
| CKAP5 | 9.2535 | 9.325 | 9.5417 | 8.3219 | 8.3264 | 8.1133 | 8.3982 |
| FANCI | 9.5455 | 8.7111 | 8.8484 | 7.2514 | 6.5865 | 5.6601 | 8.7945 |
| DNAJC9 | 11.0395 | 10.4642 | 9.6119 | 8.8527 | 8.4263 | 8.2815 | 9.8303 |
| TEX30 | 6.8132 | 6.5112 | 6.6134 | 5.575 | 4.6591 | 5.3573 | 6.1388 |
| MLC1 | 5.8958 | 5.8335 | 5.6607 | 4.6153 | 4.304 | 4.835 | 4.2499 |
| ELOVL2 | 6.0323 | 5.4283 | 5.924 | 5.1485 | 4.8761 | 4.3983 | 4.224 |
| TUBB4B | 11.9255 | 13.3122 | 12.426 | 10.7552 | 10.6633 | 11.6178 | 10.4328 |
| PARP2 | 9.359 | 8.973 | 9.4505 | 8.2942 | 7.8879 | 5.9334 | 6.586 |
| GGCT | 12.3424 | 13.2253 | 12.2131 | 11.8798 | 10.6976 | 10.1892 | 10.8668 |
| B2M | 10.6815 | 10.843 | 11.2456 | 10.1315 | 9.7091 | 9.8269 | 9.6395 |
| YKT6 | 8.6601 | 8.4224 | 8.7224 | 6.322 | 5.9566 | 6.1369 | 7.2484 |
| SYNCRIP | 9.7452 | 10.0943 | 10.6565 | 9.0701 | 9.5374 | 9.0433 | 8.42 |
| PRC1 | 9.1176 | 9.1242 | 9.9343 | 6.9378 | 6.1498 | 6.7501 | 7.783 |
| TEX2 | 8.8786 | 9.1685 | 8.3798 | 8.1639 | 7.8135 | 7.7644 | 7.972 |
| UBR7 | 9.7734 | 9.8665 | 10.1922 | 8.1243 | 7.799 | 7.5492 | 9.0737 |
| ASF1B | 9.4417 | 9.6325 | 10.4296 | 7.4971 | 8.0071 | 7.5793 | 8.9057 |
KLF10 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: KLF10
[1] KLF10 as a Tumor Suppressor Gene and Its TGF-beta Signaling.
[2] A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction.
[3] KLF10 Deficiency in CD4(+) T Cells Triggers Obesity, Insulin Resistance, and Fatty Liver.
[4] KLF10 is a modulatory factor of chondrocyte hypertrophy in developing skeleton.
[6] KLF10 Gene Expression Modulates Fibrosis in Dystrophic Skeletal Muscle.
[8] KLF10 transcription factor regulates hepatic glucose metabolism in mice.
[9] Deletion of KLF10 Leads to Stress-Induced Liver Fibrosis upon High Sucrose Feeding.
[10] FBW7 targets KLF10 for ubiquitin-dependent degradation.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below