BRD4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN68
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: radicicol
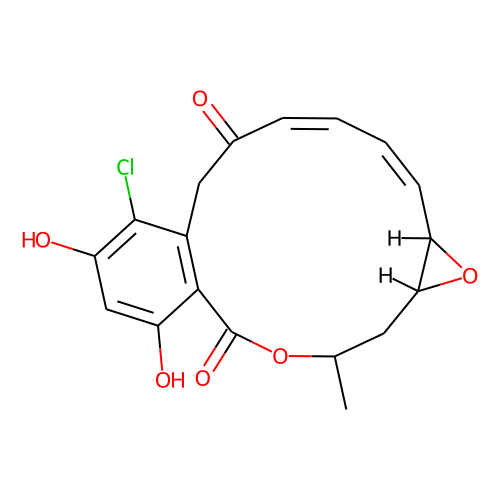
| ID | BRD-A39996500 |
| name | radicicol |
| type | Small molecule |
| molecular formula | C18H17ClO6 |
| molecular weight | 364.8 |
| condition dose | 10 uM |
| condition time | 6 h |
| smiles | C[C@@H]1C[C@@H]2[C@H](O2)/C=CC=CC(=O)CC3=C(C(=CC(=C3Cl)O)O)C(=O)O1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: BRD4
Transcription regulatory factors corresponding to this network
ARTICLE: radicicol
[2] Role of connexin 43 in cardiovascular diseases.
[3] Heat shock protein 90 inhibitors suppress pyroptosis in THP-1 cells.
[5] Development of radicicol analogues.
[7] Hydrating for resistance to radicicol.
[8] Novobiocin Analogs as Potential Anticancer Agents.
[9] Identification of Natural Compound Radicicol as a Potent FTO Inhibitor.
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition | condition |
|---|---|---|---|---|---|---|---|
| PTGDS | 10.1589 | 9.7563 | 10.1801 | 7.7557 | 8.6736 | 9.1677 | 8.9742 |
| GTF2H2B | 5.7922 | 5.3573 | 5.8077 | 5.0879 | 4.7733 | 4.9233 | 5.231 |
| CRNN | 5.0317 | 5.2956 | 5.1928 | 4.1405 | 4.6783 | 4.4579 | 4.4092 |
| EED | 9.3443 | 9.6249 | 8.7528 | 7.3769 | 7.9929 | 7.8295 | 7.9853 |
| RASA1 | 6.8521 | 7.0575 | 6.529 | 5.9217 | 6.4116 | 5.8197 | 6.2831 |
| NFKBIA | 8.8866 | 9.4157 | 9.2189 | 7.799 | 6.8763 | 6.4167 | 7.0914 |
| SOX4 | 8.3368 | 8.2142 | 8.5518 | 7.1588 | 7.0919 | 7.3268 | 7.3625 |
| MSH6 | 9.9111 | 10.0575 | 10.1468 | 8.2225 | 9.5914 | 9.3106 | 8.5312 |
| PRKCD | 8.374 | 8.4503 | 8.4258 | 7.8222 | 6.9874 | 7.4142 | 6.3449 |
| NUP62 | 8.3251 | 8.7004 | 7.7943 | 6.9721 | 7.1197 | 7.1057 | 7.5756 |
| B4GAT1 | 7.6858 | 7.9762 | 7.0339 | 6.9336 | 6.0248 | 5.842 | 6.0321 |
| MAP3K4 | 5.2359 | 5.4872 | 5.0875 | 4.6495 | 4.832 | 4.4704 | 4.3893 |
| PIP4K2B | 4.3682 | 4.8992 | 4.868 | 3.9671 | 4.0391 | 3.8626 | 4.2122 |
| CRTAP | 6.7066 | 6.8681 | 7.0928 | 5.7058 | 5.55 | 5.2871 | 6.2368 |
| CSK | 10.3925 | 10.1352 | 10.5962 | 9.7827 | 9.6411 | 8.8939 | 9.4629 |
| DDB2 | 9.166 | 9.9993 | 9.8981 | 7.7884 | 9.0215 | 8.1345 | 8.9951 |
| BAMBI | 11.8258 | 11.9684 | 11.6303 | 9.4101 | 10.5633 | 9.7543 | 10.6379 |
| SLC5A6 | 9.9369 | 9.5744 | 10.2004 | 8.4305 | 8.5761 | 8.4982 | 8.8467 |
| TBX2 | 9.354 | 9.9677 | 9.6627 | 8.4902 | 8.3471 | 6.9248 | 8.5673 |
| TCFL5 | 5.7301 | 5.6664 | 5.6198 | 3.1614 | 5.1612 | 4.0135 | 4.4091 |
| TSEN2 | 8.0319 | 7.5363 | 7.8582 | 6.3725 | 6.7271 | 7.294 | 6.8753 |
| CDCA4 | 8.3489 | 9.1602 | 8.8205 | 7.4345 | 7.167 | 6.9248 | 7.0514 |
| RRS1 | 7.8675 | 8.2954 | 8.3419 | 6.3232 | 7.2111 | 7.1146 | 7.1859 |
| RPS6KA1 | 9.6171 | 9.8035 | 9.6479 | 8.8768 | 8.7722 | 8.8462 | 8.8865 |
| ID2 | 4.9528 | 5.6878 | 5.0714 | 3.8447 | 4.5739 | 4.5426 | 4.452 |
| TLE1 | 9.262 | 9.4762 | 9.1958 | 8.7874 | 8.8843 | 8.2431 | 8.5673 |
| GATA3 | 10.0087 | 9.7897 | 9.9718 | 9.5161 | 9.0037 | 8.6878 | 9.0276 |
| CTTN | 7.8029 | 7.255 | 8.1017 | 3.9053 | 6.0349 | 5.6209 | 6.5417 |
| TSPAN4 | 10.3271 | 10.6095 | 10.6112 | 9.9749 | 9.9822 | 9.6512 | 9.9394 |
| JADE2 | 6.1198 | 6.6854 | 6.557 | 5.4376 | 5.4378 | 5.0221 | 5.4797 |
| SNX11 | 7.6245 | 6.8611 | 6.9083 | 5.5359 | 5.8412 | 6.465 | 5.9468 |
| FADD | 9.4787 | 9.3032 | 9.8803 | 8.9976 | 8.5593 | 8.1969 | 9.0373 |
| ZBTB14 | 7.138 | 7.2372 | 7.1639 | 6.1579 | 6.6621 | 6.4491 | 6.6019 |
| ERLIN1 | 7.5224 | 8.1967 | 7.8786 | 6.9509 | 7.2174 | 7.143 | 7.3352 |
| SCAF8 | 6.9143 | 7.4813 | 6.9588 | 6.516 | 6.3586 | 6.3616 | 6.5093 |
| ZC3H4 | 7.1184 | 7.1605 | 6.9625 | 6.0178 | 6.1894 | 6.1979 | 6.6126 |
| ZNF768 | 9.0347 | 9.0169 | 9.4035 | 8.1966 | 8.6329 | 8.6966 | 8.4755 |
| KAT7 | 7.2436 | 7.2583 | 7.3935 | 6.4705 | 6.6672 | 6.0451 | 6.7093 |
| ZPR1 | 7.8851 | 7.9198 | 8.3013 | 7.392 | 7.5441 | 7.2285 | 7.3722 |
| MARCKSL1 | 10.4522 | 10.0253 | 9.2684 | 7.4833 | 9.1262 | 7.7151 | 7.9096 |
| PHB | 12.9582 | 12.7654 | 12.1792 | 11.462 | 11.6323 | 11.1237 | 11.7519 |
| FAM120A | 8.4141 | 8.9473 | 8.5388 | 7.1085 | 8.075 | 7.9224 | 7.6497 |
| S100A10 | 10.3067 | 10.4513 | 10.2788 | 9.4021 | 9.4597 | 9.2584 | 9.3609 |
| SYNGR2 | 13.5757 | 12.6165 | 12.4864 | 12.2011 | 11.6244 | 11.9545 | 11.5072 |
| AMD1 | 8.3654 | 9.0733 | 8.1151 | 6.9151 | 7.9255 | 7.1513 | 6.9166 |
| IFITM2 | 7.7196 | 7.7328 | 6.5608 | 6.0695 | 6.043 | 5.0787 | 6.618 |
| HNRNPF | 10.2565 | 9.927 | 9.3951 | 9.1169 | 8.8086 | 9.0036 | 8.9851 |
| GRSF1 | 8.8039 | 8.3654 | 7.9563 | 7.0403 | 7.4969 | 7.313 | 7.5864 |
| GLB1 | 8.3465 | 8.7688 | 8.4269 | 7.2626 | 7.8436 | 7.6438 | 7.4812 |
| SRSF1 | 7.3983 | 7.5619 | 7.3644 | 6.3765 | 6.214 | 6.7064 | 6.3351 |
| ABCE1 | 8.9849 | 9.0054 | 8.817 | 8.089 | 8.198 | 8.0453 | 8.3862 |
| GSPT1 | 10.2044 | 9.7397 | 10.4439 | 9.3713 | 9.3895 | 8.7217 | 9.059 |
| LSM3 | 10.5939 | 10.8553 | 10.9793 | 10.3057 | 10.3705 | 10.109 | 9.9576 |
| BMI1 | 6.6278 | 6.969 | 6.923 | 5.2519 | 6.0651 | 6.2909 | 5.4622 |
| ITGAV | 5.9264 | 6.0961 | 5.6765 | 4.5157 | 3.8789 | 4.4444 | 5.4857 |
| NOTCH2 | 6.0636 | 6.3982 | 7.4679 | 4.6195 | 5.1893 | 5.3394 | 5.3252 |
| MBD2 | 10.057 | 9.7652 | 9.1375 | 8.0605 | 8.2672 | 8.5136 | 8.8971 |
| AFG3L2 | 8.6938 | 8.7387 | 8.679 | 7.9806 | 8.1399 | 7.4126 | 7.9286 |
| DHFR | 10.0244 | 9.7843 | 9.926 | 9.0834 | 9.5868 | 9.4295 | 9.086 |
| TYMS | 14.0679 | 13.9439 | 13.4669 | 13.0495 | 12.4426 | 12.1281 | 11.9918 |
| RALBP1 | 6.74 | 6.999 | 6.5518 | 5.8198 | 6.0749 | 5.6917 | 6.0176 |
| TRAF4 | 8.6742 | 8.7014 | 8.4786 | 7.7535 | 8.1224 | 7.6426 | 8.2456 |
| HLTF | 6.9792 | 7.8014 | 7.0913 | 6.0187 | 6.5638 | 6.2518 | 6.0469 |
| SEMA3B | 5.7804 | 6.0585 | 5.9261 | 4.9578 | 5.3679 | 4.6065 | 4.9735 |
| MYO6 | 10.821 | 9.52 | 9.7316 | 9.1927 | 9.001 | 8.7834 | 8.9034 |
| RFK | 8.8015 | 9.0151 | 8.3551 | 7.325 | 8.2162 | 7.4153 | 7.133 |
| PHYH | 8.7309 | 9.3945 | 9.693 | 8.2463 | 8.7205 | 7.6398 | 7.9285 |
| PCF11 | 6.0715 | 6.2659 | 6.65 | 5.7298 | 5.6648 | 5.8835 | 5.5972 |
| TBC1D4 | 5.8949 | 5.6699 | 5.951 | 4.967 | 4.6301 | 4.7529 | 5.1179 |
| FAM8A1 | 6.8388 | 6.3524 | 6.6116 | 6.065 | 6.1897 | 5.7269 | 5.613 |
| TMEM11 | 9.2482 | 9.5342 | 9.5341 | 8.8441 | 8.8793 | 8.9829 | 8.7054 |
| CAMLG | 9.2336 | 9.115 | 9.5603 | 8.0523 | 8.6896 | 8.0719 | 8.8092 |
| ZNF185 | 5.8314 | 7.2019 | 7.3427 | 4.5157 | 5.9967 | 5.3363 | 5.1639 |
| ST3GAL4 | 8.367 | 7.9474 | 8.0532 | 7.1666 | 7.1744 | 7.0688 | 7.3207 |
| ADARB1 | 5.1064 | 5.2164 | 4.5572 | 4.1238 | 4.0135 | 3.9824 | 4.3438 |
| USP46 | 6.3887 | 6.6511 | 6.3976 | 5.3391 | 5.8221 | 6.036 | 5.9582 |
| TSC22D2 | 9.7407 | 9.4021 | 9.8479 | 8.8182 | 8.3236 | 8.0275 | 8.4701 |
| POLA2 | 7.0368 | 7.661 | 7.8492 | 6.8204 | 6.8172 | 6.6137 | 6.7694 |
| FKBP5 | 7.3745 | 7.3119 | 6.9101 | 6.4425 | 6.4071 | 5.6067 | 6.1171 |
| SELL | 1.7687 | 1.5443 | 2.4661 | 0 | 0.6973 | 0.6587 | 0.9437 |
| MAL | 0.9712 | 0.8036 | 1.5566 | 0 | 0.099 | 0 | 0.2488 |
| CPS1 | 6.6123 | 6.7216 | 6.8795 | 5.6704 | 5.5551 | 4.3843 | 5.9591 |
| TFF1 | 13.4966 | 13.6792 | 14.6519 | 9.016 | 11.8957 | 9.1386 | 8.6193 |
| SCO2 | 9.2717 | 9.158 | 8.4525 | 8.066 | 8.3923 | 7.7457 | 8.1191 |
| TRAF1 | 5.358 | 5.301 | 5.5814 | 4.1333 | 4.6128 | 4.7046 | 4.4455 |
| BATF | 6.7875 | 6.7341 | 6.178 | 6.224 | 5.3579 | 5.3179 | 5.3459 |
| SPINK2 | 7.2677 | 5.4483 | 5.8089 | 4.3855 | 4.2349 | 4.3705 | 5.014 |
| HOXC6 | 6.8749 | 4.5733 | 5.3616 | 3.4548 | 4.0046 | 3.5984 | 2.704 |
| ARHGAP12 | 6.2311 | 6.6882 | 6.586 | 5.5495 | 5.5191 | 5.7641 | 6.0827 |
| PA2G4 | 13.0758 | 12.6116 | 12.7316 | 11.4059 | 12.1306 | 11.8998 | 12.2239 |
| APLP2 | 6.7432 | 5.9453 | 5.8959 | 5.301 | 4.9174 | 4.5178 | 5.6082 |
| XPO1 | 10.6244 | 10.6218 | 10.2444 | 8.8464 | 9.7529 | 9.83 | 9.774 |
| MRPL3 | 10.7815 | 10.7787 | 10.6797 | 10.1203 | 10.0532 | 10.258 | 9.9702 |
| CCNG1 | 7.7059 | 7.3042 | 7.6576 | 5.6493 | 6.257 | 6.5234 | 6.3777 |
| C1QBP | 14.2594 | 14.4543 | 14.3697 | 13.5636 | 13.8018 | 13.3978 | 13.225 |
| CLNS1A | 10.0775 | 9.4153 | 9.063 | 8.3674 | 8.0137 | 8.7173 | 8.6457 |
| TAX1BP3 | 15 | 14.7984 | 14.8807 | 14.3237 | 14.3237 | 13.646 | 13.9753 |
| HIGD2A | 12.3661 | 12.6689 | 13.2014 | 12.0684 | 11.5937 | 12.0652 | 12.0496 |
| XPC | 7.1485 | 7.3748 | 7.878 | 6.4365 | 7.0204 | 6.7256 | 6.7251 |
| UTP3 | 8.8639 | 8.8471 | 8.9935 | 7.2995 | 7.8379 | 8.2216 | 8.0826 |
| TOR1B | 8.3831 | 8.2686 | 8.266 | 7.3445 | 7.9191 | 7.8051 | 7.7468 |
| SLC19A2 | 7.1829 | 7.5516 | 7.398 | 6.3153 | 6.5673 | 6.7556 | 6.6829 |
| SERPINB9 | 7.3506 | 6.5141 | 6.9719 | 5.8619 | 5.4233 | 5.6604 | 6.2738 |
| GPD2 | 7.1282 | 7.0026 | 6.79 | 6.2926 | 6.2527 | 6.0418 | 6.6753 |
| PARD3 | 8.2721 | 8.0499 | 8.2301 | 7.9422 | 7.3754 | 7.2359 | 7.0193 |
| TNFSF13 | 7.6607 | 6.838 | 6.8413 | 5.7087 | 5.755 | 6.1206 | 5.8378 |
| NCOA4 | 6.6153 | 6.3013 | 6.3084 | 5.8147 | 5.521 | 5.2559 | 5.4632 |
| SERPINB6 | 11.28 | 9.7179 | 10.2077 | 9.1588 | 9.649 | 8.2228 | 8.2195 |
| SLC19A1 | 6.8816 | 7.1288 | 6.7384 | 6.2811 | 6.0133 | 6.1478 | 6.4126 |
| KPNA2 | 14.7984 | 14.6519 | 13.8216 | 12.4027 | 13.1781 | 13.4253 | 13.7725 |
| MCM3AP | 7.0645 | 7.1411 | 7.409 | 6.3622 | 6.7612 | 6.4632 | 6.806 |
| TBC1D1 | 6.2703 | 6.5934 | 6.3725 | 5.8262 | 5.6238 | 5.2924 | 5.8145 |
| UBE2E1 | 8.2648 | 8.7582 | 9.0121 | 6.6969 | 6.8459 | 7.8764 | 7.7677 |
| HACD2 | 8.8438 | 9.0626 | 8.4931 | 5.6264 | 7.7425 | 7.5194 | 7.558 |
| PIP4K2A | 6.3219 | 6.4442 | 6.2276 | 4.5001 | 5.8321 | 5.1711 | 5.4465 |
| TRIM37 | 13.6683 | 13.8018 | 13.3079 | 11.381 | 12.8158 | 12.5982 | 12.1947 |
| CDR2L | 8.8364 | 8.967 | 8.6741 | 7.6406 | 8.3711 | 7.6816 | 8.2021 |
| PFAS | 9.0054 | 9.4465 | 9.6148 | 7.9764 | 8.6219 | 8.9239 | 8.4493 |
| APRT | 12.4268 | 13.0129 | 13.4537 | 11.9179 | 12.0167 | 11.9777 | 12.3661 |
| NOP16 | 11.9576 | 12.4521 | 12.5579 | 11.5466 | 11.2641 | 11.1803 | 11.7181 |
| CTSW | 5.9161 | 5.7831 | 5.7867 | 4.4701 | 4.4212 | 4.333 | 4.4015 |
| WDR43 | 9.9142 | 9.9369 | 10.1573 | 9.0112 | 9.1495 | 8.2954 | 9.5468 |
| TTC28 | 5.4777 | 5.1186 | 5.7261 | 4.6301 | 4.77 | 4.8526 | 4.6558 |
| DUSP10 | 4.8902 | 4.9244 | 5.69 | 4.374 | 3.7211 | 3.634 | 4.3618 |
| PDCD6IP | 8.5905 | 8.7757 | 9.0982 | 7.6873 | 7.9086 | 8.3976 | 7.9429 |
| UBE2Z | 9.1064 | 9.3442 | 9.2608 | 8.1945 | 8.6645 | 8.5146 | 8.9144 |
| TMEM43 | 6.6061 | 6.8004 | 6.9347 | 6.042 | 6.507 | 5.8935 | 6.0803 |
| POLDIP2 | 9.962 | 9.955 | 9.576 | 8.9644 | 9.262 | 9.088 | 9.0702 |
| MAPKAP1 | 8.0548 | 8.2837 | 8.4565 | 7.6816 | 7.8049 | 7.4432 | 7.698 |
| YTHDF2 | 7.8328 | 8.0354 | 8.1387 | 7.0298 | 7.3825 | 6.9211 | 7.1495 |
| SYNCRIP | 9.637 | 9.904 | 9.6888 | 8.7149 | 9.3385 | 8.526 | 9.1873 |
| PPA1 | 12.8437 | 12.0186 | 10.9501 | 10.0815 | 10.1555 | 10.1564 | 10.7367 |
| THOC7 | 13.2014 | 13.5636 | 13.9886 | 12.2389 | 13.21 | 12.158 | 12.6628 |
| DAZAP1 | 9.1292 | 9.3178 | 9.5625 | 8.5524 | 8.8129 | 8.587 | 8.8322 |
| POMT1 | 6.6762 | 6.6831 | 6.6769 | 5.8697 | 6.093 | 6.0335 | 5.7641 |
| RAP2C | 9.472 | 9.7364 | 9.556 | 8.8244 | 9.2853 | 8.753 | 8.7484 |
| FASTKD1 | 7.3558 | 7.4402 | 7.3126 | 6.7344 | 6.9339 | 6.5377 | 6.6875 |
| PHF10 | 8.5636 | 9.086 | 8.473 | 7.0443 | 7.7351 | 7.4645 | 8.3213 |
| GTF3C4 | 6.754 | 6.3275 | 6.6153 | 5.7245 | 6.2092 | 5.813 | 5.572 |
| ZDHHC14 | 6.2141 | 6.6019 | 5.8855 | 4.7723 | 5.4425 | 5.5512 | 5.2334 |
| PRR11 | 9.8924 | 10.1365 | 9.6483 | 8.9425 | 8.8893 | 9.0135 | 9.3748 |
| COA7 | 8.6054 | 8.7482 | 8.5685 | 7.7368 | 8.2625 | 8.0145 | 7.9855 |
| GTDC1 | 5.9418 | 5.8936 | 5.8171 | 5.24 | 5.5057 | 4.927 | 5.3333 |
| SLIRP | 12.7179 | 12.8342 | 13.0859 | 11.6838 | 12.6289 | 11.7279 | 12.0417 |
| CHPT1 | 8.3001 | 9.2702 | 8.2463 | 6.8074 | 7.1839 | 7.2353 | 7.138 |
| IL27RA | 7.2476 | 7.3046 | 6.4901 | 5.9396 | 5.9219 | 6.2238 | 6.4235 |
| TNIP2 | 9.4282 | 8.9632 | 9.3281 | 8.1971 | 8.4791 | 8.5467 | 8.5247 |
| OXLD1 | 11.1183 | 10.799 | 11.2073 | 10.2055 | 10.0518 | 10.2484 | 10.3264 |
| HSD11B2 | 5.4961 | 5.717 | 5.5365 | 3.899 | 5.3644 | 4.3003 | 4.2401 |
BRD4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network