EGR1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
NETWORK ID: CN7356
In order to ensure the specificity of data, an ID index is built by the database itself, and each result corresponds to an ID.
CONDITION: SAR-245409
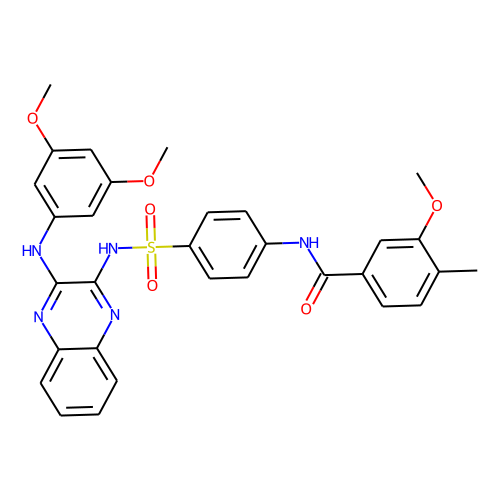
| ID | BRD-K75308783 |
| name | SAR-245409 |
| type | Small molecule |
| molecular formula | NA |
| molecular weight | NA |
| condition dose | 10.0 um |
| condition time | 24 h |
| smiles | COc1cc(Nc2nc3ccccc3nc2NS(=O)(=O)c2ccc(NC(=O)c3ccc(C)c(OC)c3)cc2)cc(OC)c1 |
CONDITION TYPE: trt_cp
Compound
CELL LINE: MCF7
Cell lines used for experiments
TRANSCRIPTION FACTOR: EGR1
Transcription regulatory factors corresponding to this network
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | KEGG pathway |
|
MTOR 
|
hsa04930 
|
|
PIK3CG 
|
hsa00562 
|
Differential expressed genes in this transcriptional regulatory sub-network
| logfc | > 0.45 and P < = 1e-5. If you want to view or download the data, you can click [expand]
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| CDC42SE1 | 8.5508 | 8.8608 | 9.112 | 9.5271 | 9.7546 | 9.9551 |
| SPATA20 | 8.3253 | 7.8744 | 8.758 | 9.5989 | 9.8437 | 9.9571 |
| RAB25 | 8.0091 | 8.7742 | 8.1365 | 9.0566 | 9.3286 | 9.2085 |
| TSKU | 6.5659 | 7.1666 | 6.929 | 7.7946 | 7.619 | 7.8414 |
| AP1M2 | 10.2871 | 10.6546 | 10.413 | 11.3583 | 11.6706 | 11.0405 |
| ZC3H7A | 8.0717 | 8.1906 | 8.0972 | 8.7513 | 8.9281 | 8.7317 |
| HEBP1 | 8.1035 | 7.7447 | 10.137 | 10.1129 | 10.3274 | 10.3566 |
| DPP3 | 9.8758 | 10.0621 | 10.3185 | 10.4864 | 10.8641 | 11.3103 |
| S100A14 | 9.9353 | 10.9667 | 10.9255 | 11.9211 | 12.0202 | 11.1897 |
| C14orf132 | 9.9992 | 10.0492 | 10.1914 | 10.9074 | 10.8808 | 11.4843 |
| SPRR3 | 2.2691 | 2.8284 | 2.5616 | 3.3789 | 4.4927 | 3.8719 |
| RHOF | 3.5391 | 3.486 | 4.7928 | 5.0734 | 4.8679 | 5.2511 |
| AMDHD2 | 9.0555 | 8.5536 | 9.2937 | 9.2075 | 11.5083 | 11.7018 |
| OGDHL | 5.6196 | 5.051 | 5.94 | 7.1312 | 6.2879 | 6.3284 |
| ESRP2 | 7.9692 | 7.9345 | 8.5667 | 9.4905 | 8.9979 | 8.6846 |
| ACSS3 | 5.326 | 5.5356 | 5.3636 | 6.2005 | 7.1761 | 7.0873 |
| GALNT6 | 9.5325 | 9.2649 | 9.6356 | 10.3676 | 10.8101 | 10.9077 |
| BCAS3 | 10.6817 | 10.1819 | 12.466 | 12.6944 | 12.8328 | 13.0647 |
| PER3 | 5.5198 | 5.3984 | 5.0686 | 6.329 | 5.8065 | 6.1642 |
| HSPB8 | 6.4964 | 6.6408 | 9.1828 | 10.8674 | 9.0337 | 9.2454 |
| CHPT1 | 7.7053 | 7.706 | 7.8469 | 8.5023 | 8.4449 | 8.7603 |
| DDX5 | 10.7641 | 10.4706 | 10.7562 | 12.0473 | 12.2603 | 11.3078 |
| HNRNPM | 10.1476 | 10.1126 | 10.0169 | 10.4902 | 10.7452 | 11.1142 |
| COX4I1 | 11.5466 | 11.8301 | 12.0705 | 12.7934 | 13.1753 | 12.2125 |
| HSP90B1 | 10.0047 | 9.5677 | 10.4477 | 11.2863 | 10.9746 | 10.7763 |
| LASP1 | 7.4129 | 7.5141 | 8.0012 | 8.3512 | 8.5044 | 8.6302 |
| PRKCSH | 10.633 | 10.0113 | 11.0361 | 11.5253 | 11.5235 | 11.7547 |
| IQGAP1 | 7.8994 | 8.2667 | 7.9526 | 8.5351 | 8.7865 | 8.7615 |
| TMBIM6 | 11.5277 | 11.8399 | 12.0396 | 12.3497 | 13.4507 | 13.0163 |
| MLF2 | 12.0619 | 12.2137 | 12.619 | 12.9906 | 13.6433 | 12.8542 |
| ALDOA | 6.8893 | 6.5004 | 6.3357 | 7.1412 | 9.3616 | 10.3127 |
| PPT1 | 10.5423 | 11.0525 | 10.7206 | 11.3481 | 12.89 | 12.2273 |
| CDH1 | 8.0432 | 7.9082 | 9.5543 | 11.7322 | 11.3055 | 10.9836 |
| ARCN1 | 8.0408 | 7.5157 | 8.035 | 8.5487 | 8.8301 | 8.4785 |
| ERP29 | 10.7083 | 11.0075 | 11.0437 | 11.4284 | 11.8722 | 11.7085 |
| CDIPT | 9.5151 | 9.7253 | 10.0414 | 10.5925 | 10.2244 | 10.5575 |
| ARHGEF12 | 3.525 | 4.0449 | 3.683 | 4.7066 | 5.01 | 4.4208 |
| NQO1 | 12.3831 | 11.473 | 13.1411 | 13.5931 | 13.9411 | 13.1948 |
| PPIF | 8.8308 | 8.2908 | 9.2679 | 9.6276 | 9.7617 | 9.4772 |
| CBX1 | 9.8929 | 9.9834 | 10.7761 | 11.187 | 11.1573 | 11.1216 |
| UBL3 | 5.4518 | 5.7122 | 5.7369 | 6.8304 | 6.9635 | 6.5769 |
| KDM5B | 6.8829 | 6.8216 | 6.7195 | 8.0835 | 7.7014 | 7.5992 |
| PPP1R12A | 6.9567 | 6.8816 | 7.387 | 7.9919 | 7.8337 | 8.005 |
| DDX42 | 7.8226 | 7.1347 | 7.7251 | 8.4836 | 8.2956 | 8.6198 |
| NET1 | 8.7797 | 8.6989 | 8.781 | 9.5478 | 9.7779 | 9.804 |
| GOLGA3 | 7.4411 | 7.9722 | 7.6883 | 8.6367 | 8.2816 | 8.2713 |
| TACSTD2 | 11.083 | 10.5056 | 11.3674 | 13.1244 | 13.4996 | 12.3131 |
| CBFB | 8.6919 | 8.8663 | 9.4055 | 9.7525 | 10.0652 | 10.2025 |
| IFI27 | 4.6334 | 4.8585 | 5.9057 | 5.5874 | 6.849 | 6.866 |
| TNFAIP2 | 3.6133 | 3.2917 | 2.5797 | 3.7398 | 5.529 | 5.0122 |
| APPBP2 | 10.1101 | 9.835 | 10.3236 | 13.1352 | 13.8935 | 10.6799 |
| TOB1 | 9.524 | 8.3254 | 9.5179 | 10.7429 | 12.47 | 10.1967 |
| PIEZO1 | 11.521 | 11.0748 | 10.8261 | 12.0166 | 12.0363 | 11.9306 |
| SLC37A4 | 5.2339 | 5.0839 | 6.5069 | 6.877 | 6.8729 | 7.2841 |
| NDUFS8 | 10.0831 | 9.948 | 11.0967 | 11.1558 | 11.4756 | 11.3365 |
| CLK2 | 7.9919 | 8.0612 | 8.2851 | 8.8581 | 8.9076 | 8.6677 |
| IGF1R | 9.1293 | 8.9047 | 9.4097 | 9.867 | 9.8263 | 9.8291 |
| TDG | 9.6158 | 9.4449 | 10.2451 | 10.9818 | 11.0974 | 11.2217 |
| RPS6KB2 | 7.8106 | 7.9734 | 8.2766 | 8.4116 | 9.6321 | 9.024 |
| LUC7L3 | 9.1636 | 9.8718 | 9.9391 | 11.5071 | 11.5701 | 10.0836 |
| GAB2 | 8.1891 | 8.2314 | 8.7292 | 9.3664 | 8.9052 | 9.0509 |
| MARK2 | 5.9943 | 5.7578 | 6.122 | 6.5916 | 6.8064 | 7.0338 |
| ENOSF1 | 5.5328 | 5.1749 | 5.73 | 6.5921 | 6.2097 | 6.2761 |
| NPIPA1 | 7.6095 | 8.1607 | 8.1441 | 9.4464 | 9.6342 | 8.8776 |
| CCDC85B | 8.2612 | 7.8218 | 8.5211 | 9.0667 | 9.2516 | 8.8396 |
| CLPX | 8.2692 | 7.9265 | 8.7462 | 8.8919 | 10.5041 | 10.167 |
| ALDH3B2 | 6.0319 | 5.574 | 5.383 | 6.6217 | 7.1801 | 7.0231 |
| P2RY2 | 6.2836 | 6.7408 | 6.5073 | 7.0551 | 7.4028 | 7.1689 |
| PRB3 | 3.7062 | 3.3986 | 3.781 | 4.3676 | 4.8828 | 4.0592 |
| TNFRSF1A | 9.0614 | 9.4184 | 10.6148 | 10.573 | 11.6846 | 10.9351 |
| PTP4A2 | 8.1288 | 7.8161 | 8.1533 | 8.7573 | 8.831 | 9.4638 |
| VAT1 | 9.32 | 8.8642 | 9.538 | 10.3668 | 10.1274 | 10.2602 |
| PRDX1 | 9.3902 | 9.8851 | 9.6059 | 10.6914 | 10.9172 | 10.517 |
| OXA1L | 11.1811 | 11.1422 | 12.6883 | 12.6612 | 13.2678 | 13.0298 |
| LGALS3 | 12.5087 | 12.0303 | 13.1934 | 13.2126 | 14.0805 | 13.6723 |
| KDM2A | 6.5546 | 6.3653 | 6.7031 | 7.2875 | 7.4628 | 7.2731 |
| COMMD4 | 8.1805 | 8.0807 | 9.0087 | 9.1121 | 9.6891 | 9.7763 |
| CYB561 | 9.9851 | 9.9463 | 10.7397 | 11.2908 | 11.3975 | 11.2372 |
| KIF2C | 4.5413 | 3.774 | 3.828 | 5.2645 | 5.4751 | 5.1192 |
| BTAF1 | 5.8194 | 6.2354 | 6.1105 | 6.9344 | 6.9026 | 6.7518 |
| IRX5 | 3.9846 | 2.7152 | 4.3522 | 5.6763 | 5.0972 | 5.0501 |
| JTB | 10.4039 | 10.3607 | 10.5078 | 11.5079 | 11.0441 | 11.2819 |
| TAGLN2 | 11.0158 | 11.2803 | 10.6941 | 11.6185 | 11.7605 | 11.8398 |
| CROCCP2 | 6.7178 | 7.3814 | 6.8674 | 8.3862 | 8.0659 | 7.4554 |
| RET | 7.6614 | 7.6477 | 9.6276 | 9.8856 | 10.0353 | 10.0428 |
| ANXA9 | 6.4746 | 6.6943 | 6.5941 | 7.2298 | 7.9818 | 7.1477 |
| EIF4A1 | 12.8759 | 12.4764 | 13.2798 | 13.4278 | 13.8966 | 13.6653 |
| PRRC2C | 9.8612 | 9.1908 | 9.1205 | 10.4548 | 10.6708 | 9.9338 |
| ATXN7L3B | 10.6545 | 10.2107 | 10.2871 | 10.9478 | 11.351 | 11.4958 |
| MED13L | 6.2465 | 5.9774 | 6.8487 | 7.1508 | 7.4084 | 7.0766 |
| SLC7A1 | 9.8519 | 8.9475 | 9.1296 | 10.4355 | 10.1421 | 10.7984 |
| ELF1 | 7.2738 | 6.5367 | 7.1891 | 8.1039 | 8.3684 | 8.1658 |
| PTPN11 | 8.6342 | 9.1853 | 9.036 | 10.1307 | 10.4757 | 9.9901 |
| MSL1 | 6.207 | 6.0201 | 7.0542 | 6.9959 | 8.0718 | 7.8469 |
| TRIM37 | 11.8735 | 11.7306 | 13.5374 | 13.7259 | 13.8386 | 14.0658 |
| SETD1B | 4.8077 | 4.5565 | 5.4872 | 6.0226 | 6.181 | 6.2015 |
| APRT | 11.4774 | 11.1417 | 12.5317 | 12.5532 | 12.9007 | 13.0166 |
| GAL | 8.5146 | 9.6129 | 7.98 | 10.2773 | 9.8285 | 10.3514 |
| SPATA2L | 7.4682 | 7.2025 | 7.6578 | 7.9867 | 8.7552 | 8.3531 |
| ERBB2 | 7.8861 | 8.3525 | 8.4379 | 9.3018 | 9.2089 | 9.1424 |
| SFXN3 | 6.2593 | 6.2457 | 6.6795 | 7.6275 | 7.1499 | 7.0562 |
| OSBPL9 | 6.2506 | 6.0917 | 6.3916 | 7.3055 | 6.6538 | 7.0977 |
| ARGLU1 | 8.0272 | 7.7334 | 8.3829 | 9.3901 | 8.9199 | 8.551 |
EGR1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
ARTICLE: EGR1
[3] The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders.
[4] Egr1 mediates retinal vascular dysfunction in diabetes mellitus via promoting p53 transcription.
[5] EGR1 interacts with TBX2 and functions as a tumor suppressor in rhabdomyosarcoma.
[6] Egr1 mediates the effect of insulin on leptin transcription in adipocytes.
[8] Role of PAR1-Egr1 in the Initiation of Thoracic Aortic Aneurysm in Fbln4-Deficient Mice.
Transcription factor KEGG
The KEGG pathway of the TF in this TRN is shown below