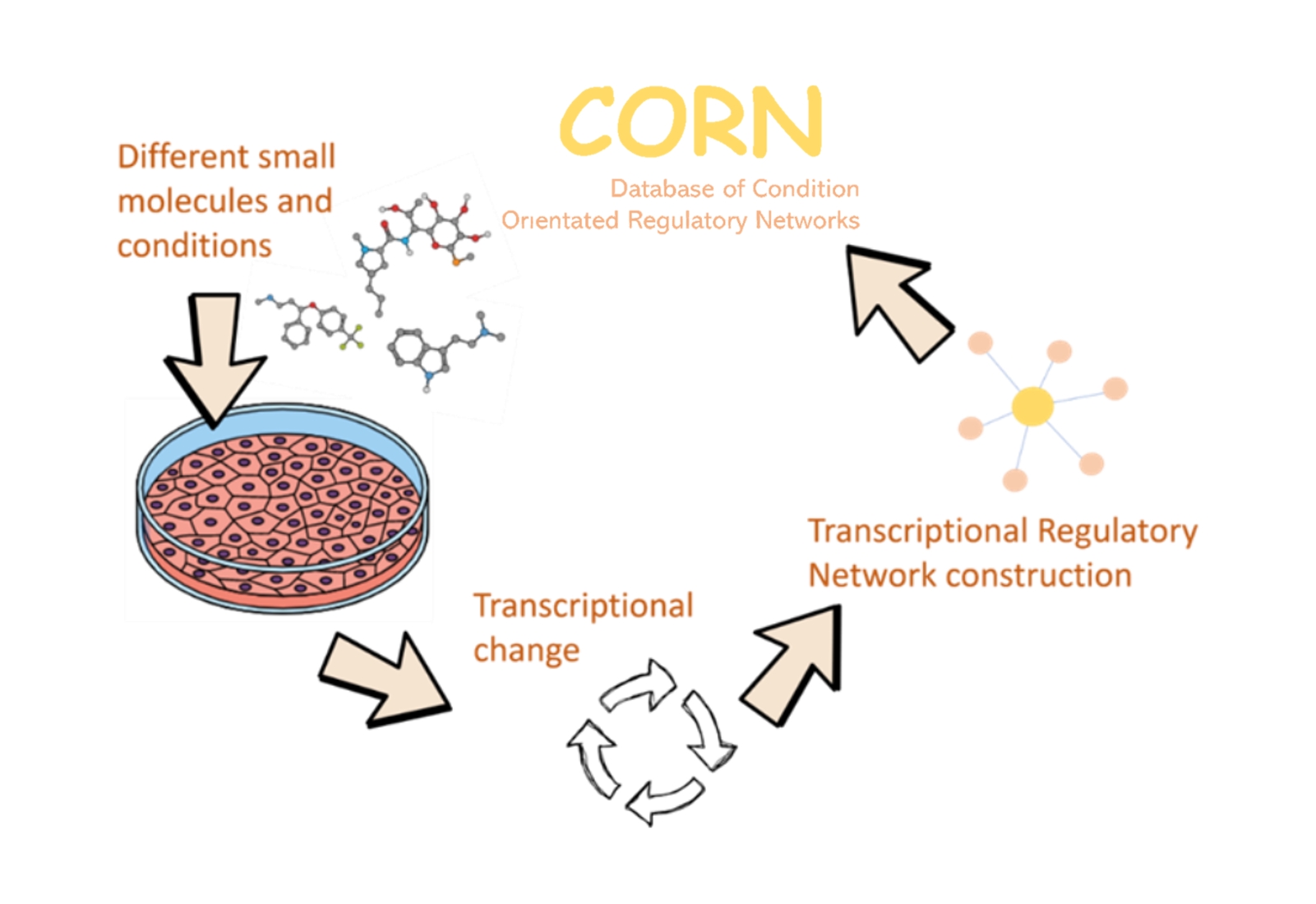
CORN is a web server designed for discovering potential treatments and transcriptional regulatory mechanisms. It hosts a comprehensive collection of condition-based transcriptional regulatory sub-networks (TRSNs). By linking and curating specific conditions to their responsive transcriptional regulatory networks (TRNs), the scientific community can now systematically understand how TRNs are altered and controlled by specific conditions in an organized manner.

CORN provides a library of condition-based TRSNs, including those induced by small molecules, drug treatments, and gene knockdowns. These TRSNs can be searched by condition, target gene, or transcription factor. The resulting condition-associated TRSN page displays detailed information, including:
- Chemical information about the drug/molecule
- The transcription factor (TF) responsible for regulating the network
- Genes regulated within the network
- The vector and fold change of such regulation
- A visual TRSN diagram
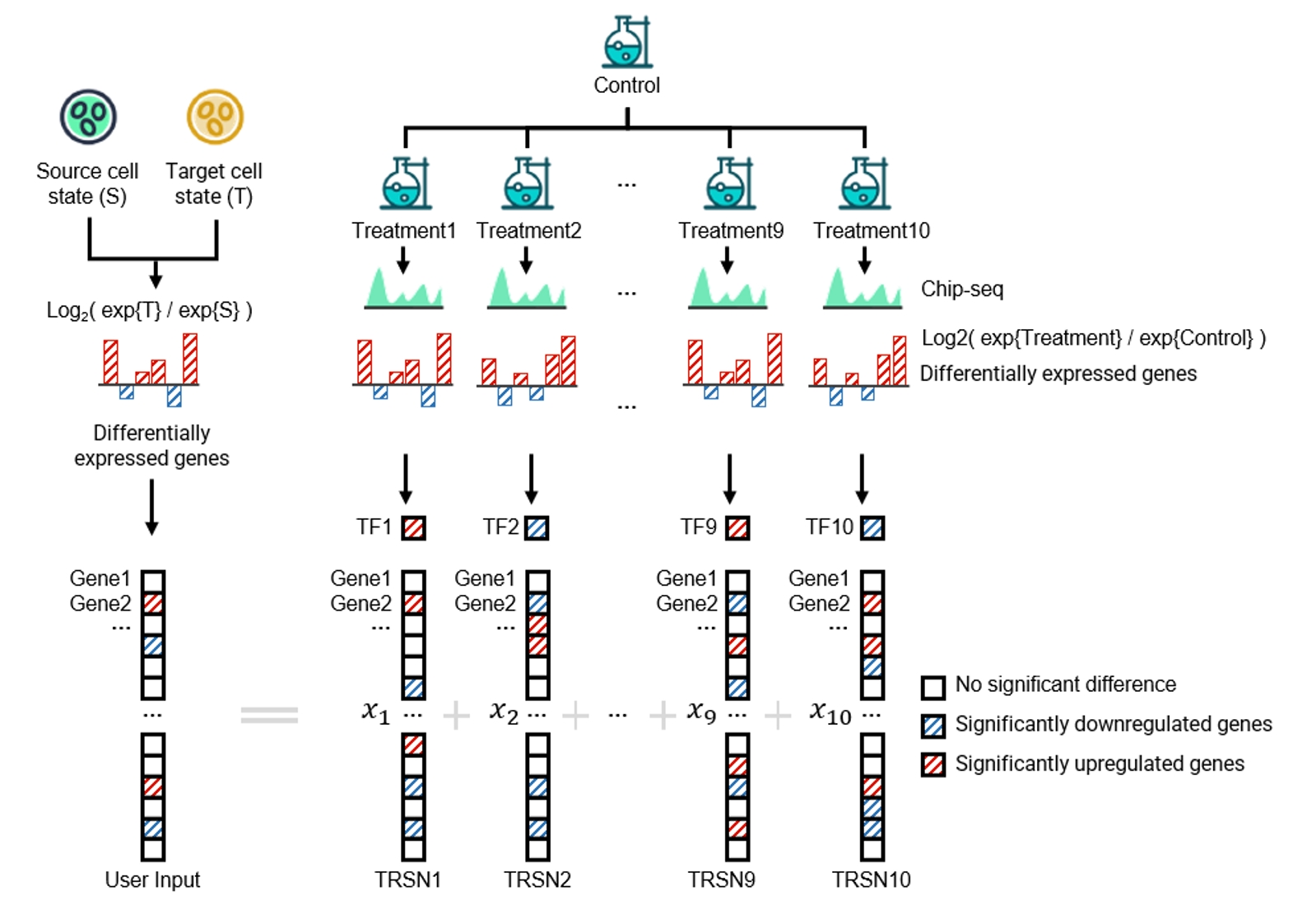
By inputting a differential expression profile of interest, our web tool matches condition-associated TRSNs with similar gene alterations using a sparse learning model. The matched TRSNs reveal the underlying gene regulatory processes, and the small molecules or drugs associated with these TRSNs can be considered potential treatments capable of modifying the input transcriptomic alterations.


