CORN HELP
This webpage provides a matching tool for discovering potential treatments and transcriptional regulatory mechanisms. It displays information on condition-based (natural compound/small molecule/drug treatments and gene perturbations) transcriptional regulatory sub-networks (TRSNs) constructed from ChIP-X data and transcriptomic changes data after specific conditional treatments in cells. Users can identify the most closely matching TRSNs using a set of input Differentially Expressed Genes (DEGs) or search and browse TRSNs by inputting the condition, target gene, or transcription factor.
Tool
Conditional Match
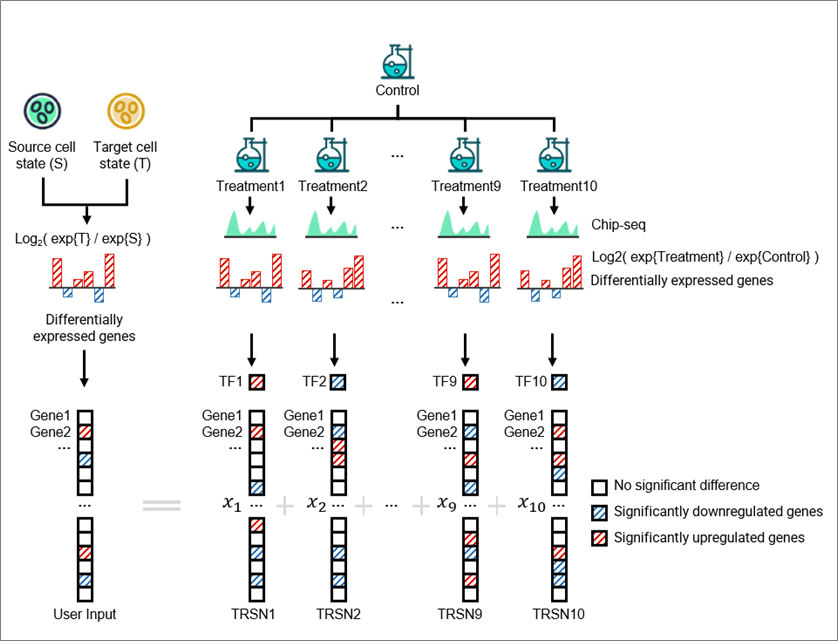
Two steps are required to perform an online conditional match:
- Select TRSN collection (Reference for matching)
- Input differentially expressed profile
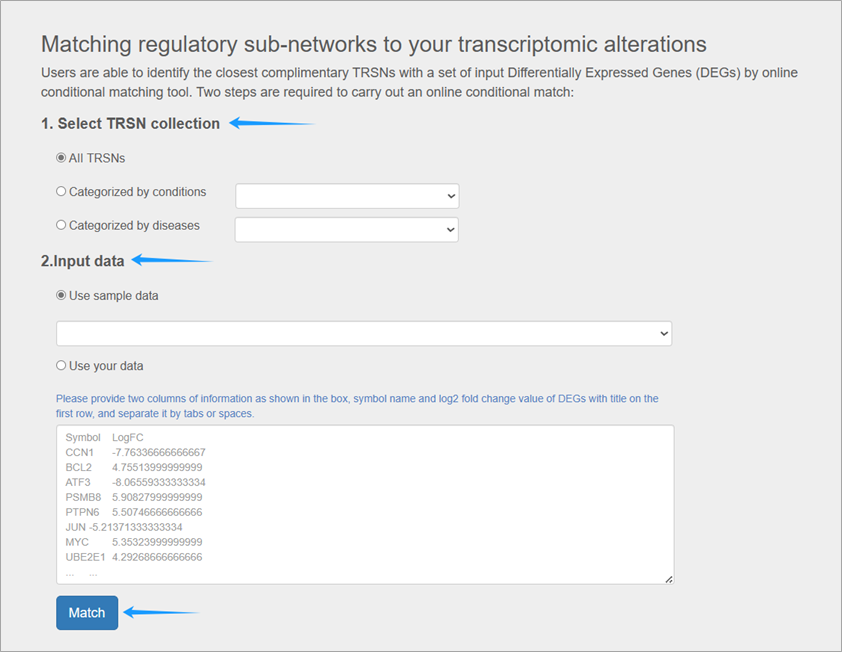
1.Select TRSN Collection (Reference for Matching)
You can choose "ALL TRSNs" or a TRSN collection of interest (categorized by condition or disease) as a matching reference. For example:
- To explore potential natural compounds, select "Categorized by conditions" → "TRSNs of natural compounds".
- To focus on breast cancer, select "Categorized by diseases" → "TRSNs of breast cancer cell lines" . Matching will then only be conducted within TRSNs from breast cancer cell lines.
2.Input Differentially Expressed Profile
We provide sample data related to applications, or you can input your own differentially expressed profile (a list of DEGs and corresponding levels of alterations in terms of log2 fold change). For example, you can input gene expression alterations between normal and cancer tissues.
3.Processing
Click "Match", and the task will run for a few minutes. The page will update automatically upon completion. Do not close this page, or you may save the result link provided.
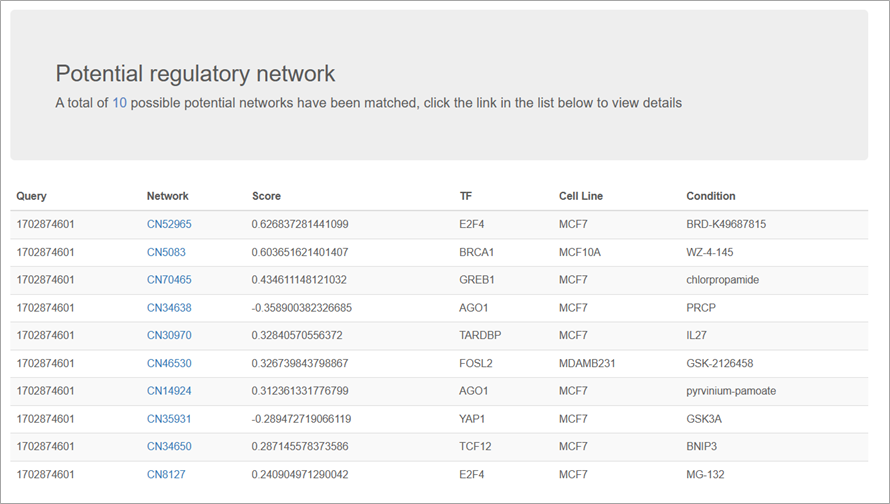
4. Results
The matching result will display the top 10 potential networks that have been matched. The "score" reflects the contribution of different TRSNs fitting into the inputted transcriptomic changes. A larger magnitude of the score indicates a closer resemblance between the TRSN and the differential expression profile.
- A negative score indicates that the regulatory direction of the inputted DEGs aligns with the regulatory direction of the reference TRSN. If researchers have a set of DEGs induced by an unknown cause, exploring matching TRSNs with negative scores would be useful.
- A positive score indicates that the inputted DEGs are in the counter-regulatory direction to the matched TRSN. Searching for condition-oriented TRSNs with positive scores would be helpful for researchers seeking conditions/drugs with therapeutic potential to counteract expression changes induced by a pathological state.
Example:
CORN RESULT1."Categorized by diseases" → "TRSNs of breast cancer cell lines"
2."Use sample data" → "Breast cancer clinical samples vs. normal samples (TCGA-BRCA)"
5.Detailed Page of TRSN
Click the Network ID (e.g., CN314) to view a detailed page describing the TRSN. The following information is included:
- Experiment information
- Condition information
- Differentially expressed genes in this TRSN
- Visualization of this TRSN
- Additional curated information about the condition and transcription factor
Information about the corresponding condition will be specified, such as the type of condition/chemical used (compound/ligand/shRNA/overexpression/CRISPR, etc.), the chemical/small molecule name, chemical structure, molecular formula, molecular weight, and simplified molecular-input line-entry system (SMILES), as well as the clinical phase.
6. Matching Tool Source Code Download
If you wish to conduct regulatory sub-network to transcriptomic alteration matching on a local server, please follow the instructions using the data and scripts downloaded from https://qinlab.sysu.edu.cn/corn/home/download_page
Search or Browse
Search by Condition
Users can search TRSNs by selecting "condition name", "condition dose", "condition time", or "cell line". The options are multi-select and allow blank entries (i.e., all-select).
Search by Target Gene
Users can search TRSNs that include certain target genes. "Target gene" is a mandatory field.
Search by Transcription Factor
Users can search TRSNs regulated by a specific transcription factor.
Browse
Users can browse the data based on the following filters: "Condition categories", "Tissue location", and "Cell line".