NETWORK ID: CN10359 |
MED1 transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | dinaciclib |
| condition dose | 2.5 uM |
| condition time | 4 h |
| CELL LINE | K562 |
| CONDITION DETALIS | ||
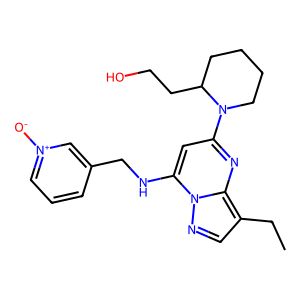 |
ID | BRD-K13662825 |
| name | dinaciclib | |
| type | trt_cp; compound | |
| molecular formula | C21H28N6O2 | |
| molecular weight | 396.5 g/mol | |
| smiles | ||
| clinical phase | Phase 3 | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| CDKN1B | 10.6807 | 12.2474 | 10.3828 | 7.929 | 7.2999 | 7.1747 |
| CDR2 | 8.2871 | 7.9303 | 7.3067 | 6.2277 | 5.8166 | 5.4712 |
| CREBBP | 8.3731 | 8.097 | 8.1434 | 6.5771 | 6.1388 | 6.6251 |
| CRKL | 11.276 | 10.6983 | 12.0296 | 8.5367 | 8.7453 | 9.1676 |
| CSTF1 | 7.4773 | 7.3708 | 7.5464 | 6.4927 | 5.9124 | 6.2539 |
| DYRK1A | 9.2612 | 9.2741 | 9.6957 | 7.6514 | 7.2317 | 7.2988 |
| E2F6 | 9.8085 | 9.4037 | 9.3444 | 7.9306 | 7.4619 | 7.9468 |
| ELF4 | 8.405 | 7.8003 | 8.4552 | 6.7852 | 6.5648 | 6.7122 |
| ERF | 6.5225 | 6.2575 | 6.1895 | 5.2589 | 5.2343 | 5.1943 |
| FOXO1 | 5.7439 | 5.478 | 5.6181 | 3.7613 | 3.9489 | 4.1375 |
| GATA2 | 9.1184 | 9.6922 | 10.6191 | 5.4885 | 4.9074 | 5.8812 |
| HES1 | 7.218 | 6.6642 | 6.6019 | 5.166 | 4.9941 | 4.5078 |
| HSF2 | 12.0745 | 11.0595 | 11.2286 | 9.8648 | 9.4843 | 9.6909 |
| DNAJB1 | 9.3403 | 9.8512 | 9.139 | 5.7883 | 5.7866 | 5.759 |
| ICAM4 | 7.5498 | 7.6463 | 7.5447 | 6.3878 | 6.5976 | 6.0753 |
| INSIG1 | 12.8694 | 12.5607 | 13.0903 | 10.2451 | 9.3086 | 10.1626 |
| LIF | 6.5215 | 6.1421 | 6.5511 | 5.0564 | 4.5558 | 4.8678 |
| MGAT2 | 10.9147 | 10.3439 | 10.706 | 8.8115 | 7.7957 | 8.6094 |
| AFF1 | 8.6842 | 8.7297 | 7.6142 | 6.364 | 6.127 | 5.9563 |
| NCK1 | 10.5224 | 10.8418 | 10.409 | 8.095 | 8.097 | 7.4475 |
| NFKBIA | 6.2506 | 6.0458 | 5.9005 | 4.8688 | 4.6274 | 4.4927 |
| CDK17 | 10.3148 | 11.018 | 10.0033 | 8.3358 | 8.876 | 7.7461 |
| PPP1R10 | 5.9966 | 6.6812 | 6.6867 | 4.8313 | 4.8876 | 4.7635 |
| PROX1 | 5.2415 | 5.4565 | 5.3262 | 4.2914 | 4.1168 | 4.4909 |
| ARID4A | 7.8656 | 7.9375 | 7.7636 | 6.0947 | 5.529 | 6.4461 |
| RNF4 | 10.0077 | 9.7288 | 10.0182 | 8.7518 | 8.0299 | 8.5604 |
| SQLE | 10.8028 | 10.8885 | 10.899 | 9.7052 | 9.7062 | 9.3369 |
| TGIF1 | 7.2908 | 6.1564 | 6.4944 | 4.0103 | 4.8629 | 4.3804 |
| YES1 | 9.5183 | 9.7729 | 9.6344 | 8.6064 | 8.4629 | 8.4979 |
| ZNF124 | 7.012 | 7.2034 | 6.9359 | 5.4507 | 6.008 | 5.7265 |
| VEZF1 | 8.7518 | 8.4869 | 8.9879 | 6.6715 | 6.877 | 7.3387 |
| ZNF200 | 7.0217 | 7.1969 | 6.9728 | 5.518 | 5.672 | 5.9348 |
| FOSL1 | 6.9402 | 6.5306 | 7.0352 | 5.1699 | 4.8963 | 4.8423 |
| PPM1D | 7.6564 | 7.9029 | 7.4943 | 5.2983 | 4.5626 | 5.1033 |
| BHLHE40 | 6.105 | 5.523 | 6.0097 | 4.4949 | 4.0601 | 3.85 |
| PER2 | 8.719 | 7.4349 | 7.7316 | 5.9653 | 5.1286 | 4.8232 |
| BCL10 | 6.7688 | 6.8589 | 6.7621 | 4.978 | 3.8784 | 5.0619 |
| SOCS6 | 7.2228 | 7.6161 | 7.62 | 6.1643 | 5.9974 | 5.6105 |
| RAB9A | 8.1019 | 8.5 | 8.5435 | 6.1946 | 6.1986 | 5.0889 |
| MED21 | 11.2945 | 11.0412 | 10.5574 | 9.595 | 9.2992 | 9.0324 |
| EIF2AK3 | 9.8449 | 10.4614 | 10.1135 | 8.5259 | 8.5066 | 8.107 |
| TBPL1 | 11.1832 | 11.2076 | 11.3948 | 9.4715 | 8.983 | 9.4002 |
| SLC25A44 | 7.0567 | 7.2791 | 6.7109 | 5.7087 | 5.5918 | 5.5164 |
| SERTAD2 | 7.1854 | 7.8456 | 7.5042 | 6.1111 | 5.155 | 4.8963 |
| URB2 | 8.4151 | 8.876 | 8.7721 | 7.3616 | 7.0735 | 7.5358 |
| KEAP1 | 9.9522 | 9.0369 | 9.6812 | 7.2839 | 6.2017 | 7.0743 |
| ZBTB39 | 6.1578 | 5.9569 | 5.884 | 4.9063 | 4.5089 | 4.5257 |
| HMGXB4 | 9.9854 | 9.8828 | 9.751 | 7.8723 | 8.5009 | 8.1964 |
| ZNF443 | 6.2804 | 5.6202 | 5.9746 | 4.4202 | 3.6318 | 3.755 |
| TIMM17A | 13.1006 | 13.2528 | 13.2441 | 12.0145 | 12.0669 | 11.4493 |
| TOB2 | 7.2754 | 7.8032 | 7.0487 | 5.2479 | 5.2751 | 5.0252 |
| CLP1 | 7.3128 | 7.2883 | 7.5133 | 6.0826 | 5.3829 | 5.5088 |
| KAT7 | 9.2188 | 9.5021 | 9.0446 | 8.0428 | 7.6753 | 7.9754 |
| MTF2 | 10.171 | 10.6191 | 10.1131 | 8.4508 | 7.8119 | 8.1178 |
| CHSY1 | 10.5779 | 9.9048 | 10.005 | 8.1432 | 7.5611 | 6.8777 |
| SEPHS2 | 10.0473 | 10.1406 | 10.107 | 8.6017 | 8.8398 | 8.9879 |
| SPEN | 10.2113 | 10.4182 | 10.9363 | 9.1551 | 8.1124 | 8.5793 |
| RRP1B | 12.632 | 12.6022 | 12.1958 | 11.0609 | 10.5833 | 11.1115 |
| ZC3H4 | 8.3049 | 8.0793 | 8.3633 | 7.0127 | 6.8672 | 7.0655 |
| SIRT1 | 7.3826 | 7.8108 | 7.2892 | 5.525 | 5.0361 | 4.9658 |
| CDC42EP4 | 6.8208 | 7.3776 | 6.5659 | 4.8912 | 4.9063 | 4.5082 |
| LEMD3 | 7.9192 | 8.4223 | 8.3505 | 6.6501 | 6.9985 | 6.5306 |
| PATZ1 | 6.7329 | 6.8306 | 6.8011 | 5.226 | 4.6414 | 5.0713 |
| NUP62 | 11.1832 | 10.6645 | 11.2841 | 9.4601 | 8.5846 | 9.2298 |
| ZNF318 | 8.6864 | 8.6217 | 8.4647 | 7.2136 | 6.6687 | 6.78 |
| AHCTF1 | 10.3439 | 10.8166 | 10.2004 | 9.198 | 9.0539 | 8.9993 |
| TIPARP | 10.0245 | 9.4657 | 9.584 | 6.1981 | 6.3955 | 6.0927 |
| NGDN | 11.4048 | 10.7738 | 10.7885 | 9.7656 | 9.2987 | 9.0928 |
| ZZZ3 | 9.511 | 9.3322 | 9.3004 | 8.391 | 7.5804 | 7.7697 |
| AGO2 | 8.521 | 8.5856 | 8.7761 | 7.3011 | 7.5563 | 7.2891 |
| PHF20 | 6.4684 | 6.8003 | 7.0022 | 5.3143 | 5.0928 | 4.8025 |
| MEX3C | 6.5902 | 6.6792 | 6.4416 | 5.3088 | 5.43 | 5.5412 |
| PCF11 | 8.1737 | 8.4 | 8.7048 | 6.97 | 6.3579 | 6.8754 |
| KLF13 | 5.747 | 5.4458 | 5.5009 | 4.4833 | 4.5013 | 4.3402 |
| MIEF1 | 8.292 | 8.8287 | 8.7783 | 7.0298 | 7.3533 | 7.3649 |
| C2orf42 | 5.9604 | 5.9458 | 5.7408 | 4.9609 | 4.7002 | 4.6001 |
| GPATCH1 | 6.9324 | 6.9308 | 6.5774 | 5.697 | 5.5449 | 5.5152 |
| DNAAF2 | 9.0697 | 8.976 | 9.5489 | 7.7705 | 7.0974 | 7.557 |
| SMG8 | 7.9271 | 7.7483 | 8.2985 | 6.0792 | 5.366 | 6.462 |
| ZNF654 | 7.3003 | 7.2567 | 7.1854 | 5.741 | 5.5914 | 5.8231 |
| ZC3HAV1 | 8.0529 | 8.2796 | 8.1777 | 7.1968 | 7.2115 | 7.1229 |
| ZBED5 | 8.4979 | 8.2838 | 8.6753 | 7.4628 | 7.0964 | 7.1635 |
| RBM15 | 10.1805 | 9.7052 | 9.9522 | 7.7614 | 6.552 | 7.4769 |
| MIS12 | 9.8153 | 9.6579 | 9.7477 | 8.6395 | 8.5264 | 8.5746 |
| PALB2 | 8.1553 | 8.2722 | 8.1964 | 6.7267 | 6.8898 | 7.0027 |
| CBLL1 | 8.6117 | 8.4482 | 8.4335 | 7.2176 | 6.8184 | 7.1539 |
| ZNF430 | 6.7645 | 6.7901 | 6.1413 | 4.5617 | 4.5542 | 4.4916 |
| PCGF1 | 7.8952 | 7.8628 | 7.7527 | 6.4288 | 6.7871 | 6.5061 |
| MFSD5 | 6.7825 | 7.0279 | 6.9491 | 5.3829 | 4.9679 | 5.0519 |
| ZNF764 | 6.5722 | 6.4983 | 6.508 | 4.7871 | 4.8547 | 4.8656 |
| TBC1D31 | 9.966 | 10.642 | 10.4327 | 8.1761 | 9.1257 | 8.4622 |
| RNF19B | 9.749 | 9.3999 | 9.4674 | 8.1025 | 8.2971 | 7.4508 |
MED1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [dinaciclib] and [MED1]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
CDK2 |
Cyclin-dependent kinase 2
|
298
|
ARTICLE: dinaciclib
[1] Dinaciclib for the treatment of breast cancer.
[5] Cyclin-dependent protein serine/threonine kinase inhibitors as anticancer drugs.
TRANSCRIPTION FACTOR: MED1
| Gene | Protein | Amino acids | More |
|
MED1 |
Mediator of RNA polymerase II transcription subunit 1
|
1581
|
ARTICLE: MED1
[1] MED1 Regulates BMP/TGF- in Endothelium: Implication for Pulmonary Hypertension.
[2] MED1 mediator subunit is a key regulator of hepatic autophagy and lipid metabolism.
[3] MED1 Deficiency in Macrophages Aggravates Isoproterenol-Induced Cardiac Fibrosis in Mice.
[4] Phosphorylated MED1 links transcription recycling and cancer growth.
[5] Med1 controls CD8 T cell maintenance through IL-7R-mediated cell survival signalling.
[8] Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer.