NETWORK ID: CN10681 |
RXRA transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | mitoxantrone |
| condition dose | 2.22 uM |
| condition time | 24 h |
| CELL LINE | MCF7 |
| CONDITION DETALIS | ||
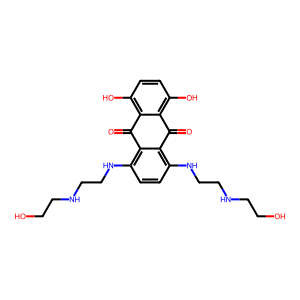 |
ID | BRD-K21680192 |
| name | mitoxantrone | |
| type | trt_cp; compound | |
| molecular formula | C22H28N4O6 | |
| molecular weight | 444.5 g/mol | |
| smiles | ||
| clinical phase | FDA Approved | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition |
|---|---|---|---|---|
| SERPINA3 | 6.7636 | 6.8345 | 6.9249 | 0.6434 |
| ALDH3B2 | 7.868 | 7.6157 | 8.0563 | 4.9314 |
| ARG1 | 4.0904 | 3.7965 | 4.1544 | 1.3133 |
| ATP6V1C1 | 8.9633 | 8.6311 | 9.0704 | 5.4861 |
| CCND1 | 8.4486 | 8.5164 | 8.6592 | 5.2815 |
| KLF5 | 7.5048 | 7.2559 | 7.1569 | 4.2789 |
| CAST | 8.4196 | 8.1034 | 8.235 | 5.5106 |
| CCNF | 8.7623 | 8.7785 | 8.7027 | 4.604 |
| CRABP1 | 11.9624 | 12.2323 | 11.9881 | 6.2423 |
| CYB561 | 9.6584 | 9.7448 | 9.7661 | 5.5106 |
| EMP2 | 9.4398 | 9.4755 | 9.726 | 6.167 |
| ESR1 | 12.3259 | 11.4947 | 11.6389 | 4.696 |
| GABPB1 | 8.7387 | 8.5971 | 8.8549 | 5.2815 |
| GATA3 | 9.4968 | 9.315 | 9.4531 | 6.2713 |
| HMGB2 | 10.4941 | 10.4156 | 10.6388 | 7.2663 |
| HSPA2 | 7.8075 | 7.59 | 7.3762 | 4.375 |
| DNAJB1 | 9.9786 | 9.8681 | 9.9793 | 7.1547 |
| ICA1 | 7.992 | 8.2283 | 8.3956 | 5.0483 |
| IMPA1 | 8.7574 | 9.0079 | 9.0567 | 5.8391 |
| KRT19 | 13.9437 | 14.4591 | 14.5055 | 9.5647 |
| LGALS3 | 13.1303 | 13.3309 | 12.6457 | 7.9199 |
| SMAD3 | 8.117 | 8.0039 | 8.2078 | 5.7647 |
| MDK | 11.0906 | 11.2486 | 11.4436 | 7.1595 |
| SCGB2A1 | 10.4335 | 10.7329 | 11.1842 | 6.7641 |
| PDGFA | 8.3247 | 8.4054 | 8.3316 | 4.604 |
| POLE2 | 9.4251 | 9.3493 | 9.2933 | 5.3572 |
| MAPK6 | 11.7077 | 12.0263 | 11.7218 | 8.9776 |
| PTK2 | 8.7168 | 8.8506 | 8.8168 | 6.0635 |
| PTPRN2 | 7.3237 | 7.4906 | 7.4957 | 4.849 |
| RAD9A | 7.3217 | 7.3825 | 7.559 | 4.8332 |
| S100A10 | 9.9627 | 10.6697 | 10.6119 | 5.2898 |
| SLC7A1 | 10.7946 | 11.0934 | 11.1975 | 7.6822 |
| SMARCE1 | 10.3356 | 10.4084 | 10.2568 | 7.6676 |
| AURKA | 7.8882 | 7.7251 | 8.1001 | 5.0165 |
| TCP1 | 9.9918 | 9.8503 | 9.7988 | 7.3188 |
| TPD52 | 8.2196 | 8.1864 | 8.6501 | 3.836 |
| XBP1 | 12.681 | 12.5325 | 12.6476 | 6.7112 |
| ZNF7 | 6.5617 | 6.6337 | 6.6222 | 3.9333 |
| IFRD2 | 10.6326 | 10.6578 | 10.7816 | 7.5123 |
| FXR1 | 8.8958 | 9.1649 | 8.7943 | 6.0515 |
| ZNF239 | 6.7729 | 6.7824 | 6.8719 | 4.4087 |
| SAP30 | 8.2225 | 8.3217 | 8.4836 | 5.3712 |
| PRPF4 | 9.1758 | 9.3431 | 9.2674 | 4.604 |
| CCNE2 | 6.9503 | 6.6502 | 6.6723 | 4.0774 |
| CXCL14 | 14.753 | 13.9986 | 14.9725 | 8.7654 |
| WTAP | 9.3264 | 9.4389 | 9.2135 | 6.6236 |
| EMC2 | 9.3778 | 9.3079 | 9.5003 | 6.7791 |
| KIAA0100 | 7.9198 | 7.9038 | 7.8276 | 5.1408 |
| DLGAP5 | 9.0127 | 9.0033 | 9.1505 | 6.4963 |
| PTDSS1 | 9.8237 | 9.7627 | 9.6688 | 6.5126 |
| TOMM20 | 8.7348 | 9.0719 | 8.9178 | 6.2912 |
| KEAP1 | 8.5168 | 8.4156 | 8.5756 | 5.1408 |
| DDX39A | 12.5014 | 12.7995 | 13.2133 | 8.8833 |
| NET1 | 9.4707 | 9.363 | 8.9281 | 5.0165 |
| LYPLA1 | 10.8581 | 10.6728 | 10.5755 | 4.3094 |
| AGR2 | 8.6645 | 8.3844 | 9.5068 | 3.1495 |
| NPRL2 | 7.3602 | 7.2309 | 7.2587 | 5.0856 |
| ARPP19 | 10.2965 | 10.5546 | 10.3848 | 5.3572 |
| MRPS30 | 9.5369 | 9.5343 | 9.6101 | 6.4021 |
| POLG2 | 8.4567 | 8.5723 | 8.3716 | 4.0774 |
| XPOT | 11.5738 | 11.9596 | 11.6272 | 8.9341 |
| EXOSC8 | 10.4332 | 10.517 | 10.514 | 8.3115 |
| DCUN1D4 | 8.2827 | 8.0437 | 7.8456 | 5.2265 |
| LEMD3 | 8.1361 | 8.1392 | 8.0714 | 5.9596 |
| ACOT9 | 8.9657 | 9.0333 | 8.6783 | 5.5106 |
| SPDEF | 12.0037 | 12.1776 | 12.1643 | 9.402 |
| MRPL15 | 11.9769 | 11.7466 | 11.8929 | 7.0551 |
| GPSM2 | 8.3518 | 8.3439 | 8.7176 | 5.1025 |
| MTERF3 | 10.9808 | 10.8581 | 10.7072 | 4.604 |
| TUBD1 | 11.3044 | 10.9403 | 11.0447 | 7.5831 |
| TRAPPC4 | 8.9633 | 9.4149 | 9.3591 | 6.0409 |
| EVL | 7.1593 | 7.7686 | 7.736 | 3.8023 |
| RAB14 | 8.2768 | 8.5585 | 8.6064 | 5.7585 |
| LSM8 | 10.8075 | 11.0144 | 11.2208 | 8.0796 |
| ESRP1 | 8.2061 | 8.0823 | 8.188 | 5.3424 |
| CDCA4 | 8.9508 | 8.6352 | 8.7577 | 5.0856 |
| UBR7 | 9.0999 | 9.0111 | 9.1796 | 5.9453 |
| ADI1 | 8.8191 | 8.7334 | 8.6993 | 4.928 |
| MCUR1 | 8.8157 | 8.6755 | 8.7108 | 5.9453 |
| ACBD3 | 7.0331 | 6.7901 | 7.1062 | 4.3962 |
| ACD | 8.8998 | 8.6553 | 8.9939 | 5.2265 |
| C8orf33 | 8.7775 | 9.5158 | 9.3168 | 4.5935 |
| TMEM223 | 9.7266 | 9.9406 | 9.7387 | 6.9884 |
| ZFAND1 | 8.5501 | 8.2997 | 8.1494 | 4.8704 |
| DENND2D | 8.8396 | 8.5538 | 8.6253 | 5.9453 |
| ASRGL1 | 8.6018 | 8.207 | 8.1443 | 4.2682 |
| TSEN2 | 8.3159 | 8.3041 | 8.479 | 5.6121 |
| PRR7 | 9.6269 | 9.6893 | 9.7138 | 7.3007 |
| CROCCP2 | 6.7667 | 6.8251 | 6.8114 | 4.2871 |
| TSPYL5 | 6.9895 | 6.7886 | 7.0217 | 4.1635 |
| MRFAP1L1 | 7.7276 | 7.9492 | 7.925 | 5.4528 |
| NEAT1 | 7.7176 | 7.8886 | 8.362 | 2.3844 |
| ATXN7L3B | 9.9702 | 10.0547 | 10.0365 | 7.8748 |
RXRA transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [mitoxantrone] and [RXRA]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
TOP2A |
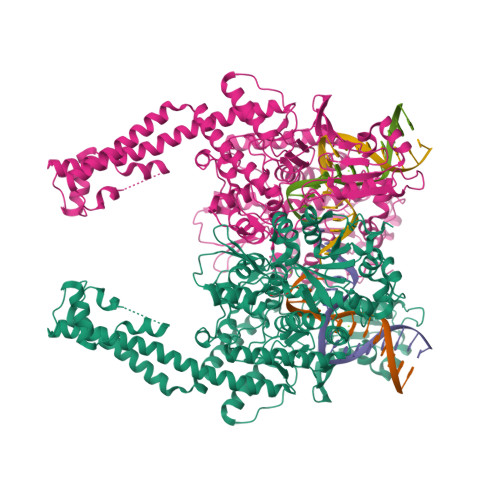
DNA topoisomerase 2-alpha
|
1531
|
ARTICLE: mitoxantrone
[1] Mitoxantrone, More than Just Another Topoisomerase II Poison.
[2] Mitoxantrone-Surfactant Interactions: A Physicochemical Overview.
[3] Mitoxantrone for multiple sclerosis.
[4] Mitoxantrone treatment of multiple sclerosis: safety considerations.
[6] Mitoxantrone: benefits and risks in multiple sclerosis patients.
TRANSCRIPTION FACTOR: RXRA
| Gene | Protein | Amino acids | More |
|
RXRA |
Retinoic acid receptor RXR-alpha
|
462
|