NETWORK ID: CN30970 |
TARDBP transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | IL27 |
| condition dose | |
| condition time | 144 h |
| CELL LINE | MCF7 |
| CONDITION DETALIS | ||
| ID | TRCN0000058478 | |
| name | IL27 | |
| type | trt_sh; shRNA | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| ADK | 11.2342 | 10.8908 | 10.7722 | 11.5963 | 11.743 | 11.8934 |
| FAS | 4.8671 | 4.6697 | 4.8503 | 7.3424 | 7.0628 | 6.8053 |
| CAPN3 | 8.1642 | 7.9496 | 8.254 | 8.7815 | 9.2793 | 9.5415 |
| CDH5 | 5.3829 | 5.2711 | 6.2109 | 6.9426 | 7.3358 | 6.9311 |
| CDKN1A | 13.4748 | 13.4089 | 13.8327 | 15 | 14.4247 | 15 |
| CDKN1C | 6.6537 | 6.3231 | 6.9513 | 7.375 | 8.3356 | 8.1918 |
| CKB | 10.2448 | 10.7271 | 10.6616 | 12.081 | 12.2361 | 12.4249 |
| DDB2 | 12.1461 | 11.814 | 10.6181 | 13.0464 | 13.2575 | 13.2248 |
| DPH2 | 8.3373 | 8.0709 | 7.7682 | 9.5381 | 10.0495 | 9.708 |
| EDN1 | 7.6384 | 7.6993 | 8.0429 | 8.7557 | 9.5166 | 9.7726 |
| ELN | 7.2824 | 6.8451 | 6.7585 | 8.6397 | 8.8898 | 8.3446 |
| FCGR2B | 5.5788 | 6.0318 | 5.4342 | 6.4706 | 6.7 | 6.5878 |
| FOLR1 | 5.1355 | 4.3736 | 5.1528 | 6.0373 | 6.585 | 7.0277 |
| GALNS | 10.4636 | 10.629 | 10.2833 | 11.7162 | 11.1803 | 11.3 |
| GALNT2 | 7.0059 | 6.909 | 7.2176 | 8.4883 | 9.8903 | 9.2584 |
| GBE1 | 6.2492 | 6.9563 | 6.5945 | 7.5167 | 8.5634 | 8.3037 |
| HOXA5 | 5.2696 | 4.9896 | 5.3098 | 5.8105 | 6.7458 | 6.7902 |
| HOXA7 | 7.0722 | 7.2933 | 7.5142 | 8.2495 | 8.1285 | 8.449 |
| IDH3A | 9.4608 | 10.1644 | 9.854 | 10.7908 | 11.1427 | 10.9344 |
| ITGB2 | 5.0695 | 5.3326 | 5.5145 | 6.2277 | 6.2938 | 6.0103 |
| L1CAM | 10.1089 | 9.4787 | 10.211 | 10.9772 | 11.038 | 11.757 |
| LEP | 4.9155 | 4.5754 | 4.621 | 5.2552 | 5.7747 | 5.6981 |
| MAK | 6.4199 | 6.1981 | 6.127 | 6.7747 | 6.8497 | 6.9324 |
| MCAM | 6.6923 | 6.3062 | 7.0001 | 8.2052 | 8.2575 | 8.7674 |
| MYD88 | 10.6442 | 10.2352 | 10.5954 | 11.0848 | 11.2758 | 11.4683 |
| MYO1E | 5.7778 | 6.2428 | 6.5062 | 7.9864 | 8.3709 | 7.5152 |
| NAGLU | 8.9384 | 9.0093 | 9.0916 | 9.8802 | 9.8905 | 10.3916 |
| NFKBIE | 8.2027 | 8.4176 | 8.0993 | 9.1272 | 9.0367 | 9.2253 |
| NPR1 | 5.5251 | 6.2555 | 6.0423 | 7.4427 | 6.9518 | 6.9167 |
| OXTR | 3.0635 | 2.997 | 2.7141 | 5.4473 | 5.8745 | 4.7199 |
| PDGFB | 6.7685 | 7.3615 | 7.6198 | 8.1918 | 8.5415 | 8.7034 |
| PHKG2 | 8.5459 | 8.4423 | 8.8353 | 10.0163 | 10.4774 | 10.5909 |
| PIK3C2B | 7.9029 | 7.485 | 7.8825 | 8.893 | 10.1149 | 9.84 |
| PLIN1 | 2.8226 | 3.6479 | 2.6421 | 4.4969 | 5.6333 | 5.549 |
| PLOD1 | 11.6348 | 11.4797 | 12.0705 | 12.8899 | 12.6542 | 12.7112 |
| PPARG | 3.5516 | 3.1648 | 4.3416 | 5.6284 | 7.045 | 6.585 |
| PPP2R5B | 6.4918 | 6.0818 | 6.0228 | 7.1077 | 7.0376 | 7.8612 |
| PRM2 | 6.3699 | 6.349 | 6.512 | 6.8697 | 7.0963 | 7.1122 |
| RPS6KA2 | 3.3077 | 2.9736 | 3.8201 | 4.9516 | 4.8583 | 6.238 |
| S100A4 | 7.7703 | 8.1222 | 6.7613 | 12.0398 | 11.7134 | 11.133 |
| SLC19A1 | 8.7296 | 8.6997 | 8.7904 | 9.2344 | 9.3177 | 9.5726 |
| SNCG | 9.285 | 9.4588 | 9.5992 | 10.2073 | 10.4176 | 10.0641 |
| FSCN1 | 9.8498 | 10.0949 | 9.683 | 11.2692 | 10.8043 | 10.7205 |
| TNFAIP2 | 4.9318 | 4.4646 | 5.5644 | 7.3711 | 6.5642 | 6.3699 |
| TNNI2 | 8.8433 | 8.9212 | 8.4468 | 9.5239 | 9.4604 | 9.9954 |
| TRPM2 | 7.4309 | 7.0809 | 7.2417 | 7.9696 | 8.0042 | 7.775 |
| PHLDA2 | 10.1276 | 10.0507 | 10.4014 | 11.0582 | 11.1083 | 10.8056 |
| TUFM | 13.8006 | 14.021 | 13.7203 | 14.5669 | 14.8001 | 14.8034 |
| TNFRSF4 | 5.9332 | 6.2563 | 6.3384 | 6.8515 | 7.1061 | 6.8853 |
| CAMK1 | 10.0413 | 10.215 | 10.3355 | 10.9765 | 10.8554 | 11.4429 |
| KCNK5 | 5.5179 | 5.5856 | 5.6302 | 6.027 | 6.3584 | 6.5956 |
| ALDH4A1 | 9.626 | 9.3242 | 8.9688 | 10.278 | 10.7722 | 10.5059 |
| EIF3B | 11.8836 | 11.9049 | 11.8163 | 13.135 | 12.4331 | 12.9163 |
| EIF4G3 | 10.9235 | 11.1614 | 10.9868 | 11.7311 | 12.1712 | 11.6802 |
| INPP4B | 10.0333 | 10.4366 | 9.5611 | 11.2787 | 11.086 | 11.4973 |
| SYNJ1 | 6.972 | 6.9437 | 7.5679 | 8.0536 | 8.1677 | 7.9819 |
| PDLIM1 | 10.5189 | 10.5392 | 10.5546 | 11.4335 | 11.4021 | 11.8587 |
| WDR46 | 9.5923 | 9.7195 | 10.0613 | 10.6401 | 10.7006 | 11.16 |
| ADIPOQ | 2.5514 | 3.4223 | 2.477 | 3.9987 | 5.2136 | 5.3081 |
| GOSR2 | 6.9928 | 7.1497 | 7.4086 | 8.0879 | 8.1408 | 8.5082 |
| AREL1 | 5.2967 | 5.6693 | 6.3116 | 6.7909 | 7.7654 | 7.5232 |
| TECPR2 | 7.209 | 7.3625 | 7.6091 | 8.3531 | 8.4346 | 7.9639 |
| AP5Z1 | 7.7429 | 8.0301 | 8.3907 | 9.053 | 9.0549 | 9.8142 |
| DGCR2 | 8.5602 | 8.078 | 8.5394 | 9.3905 | 9.182 | 9.3116 |
| KATNB1 | 7.7824 | 8.0507 | 8.0226 | 8.4856 | 8.704 | 8.61 |
| TCIRG1 | 7.4246 | 6.9374 | 8.7034 | 9.953 | 10.3955 | 10.1835 |
| CITED2 | 8.2158 | 9.1352 | 8.5358 | 9.9117 | 10.1838 | 9.8052 |
| ZNHIT1 | 10.9184 | 10.7479 | 10.941 | 11.7798 | 11.7882 | 11.9495 |
| NUDC | 11.7812 | 11.9434 | 12.1129 | 12.4948 | 12.666 | 12.599 |
| EBNA1BP2 | 12.3427 | 12.4315 | 12.2979 | 13.1763 | 12.9784 | 13.0662 |
| GLIPR1 | 1.3057 | 1.1826 | 0.4665 | 3.1216 | 2.1981 | 2.6294 |
| WWP2 | 8.4543 | 8.4887 | 8.1364 | 9.3577 | 9.3851 | 10.0919 |
| SAMD4A | 8.929 | 8.5751 | 8.1554 | 9.9146 | 10.3585 | 9.7645 |
| PLXND1 | 8.6182 | 8.4033 | 10.0676 | 11.3415 | 11.5424 | 11.5831 |
| RRP12 | 7.166 | 7.9658 | 6.8261 | 9.0196 | 9.9907 | 10.306 |
| IQCE | 7.9773 | 8.2393 | 8.2473 | 8.6967 | 9.3314 | 9.3926 |
| ATMIN | 9.2362 | 9.1971 | 9.8079 | 10.7621 | 11.4748 | 10.8894 |
| ATP13A2 | 7.0608 | 7.2283 | 7.0336 | 7.8896 | 8.148 | 8.9064 |
| MLYCD | 8.9722 | 8.9551 | 9.2537 | 9.7806 | 10.1747 | 9.7255 |
| PHLDA3 | 9.1305 | 9.0259 | 9.7279 | 10.8295 | 11.1283 | 10.9842 |
| PLA2G15 | 7.1397 | 6.6246 | 7.8046 | 8.5455 | 9.2866 | 8.739 |
| MTCH1 | 11.2147 | 11.0572 | 11.3584 | 11.7115 | 11.8803 | 11.9114 |
| MTCH2 | 11.2467 | 11.701 | 11.9803 | 12.8131 | 12.857 | 13.2139 |
| YIPF3 | 10.589 | 10.5252 | 10.8911 | 11.4403 | 11.4748 | 11.5505 |
| B3GAT3 | 9.6431 | 9.5833 | 9.4769 | 10.2833 | 10.1189 | 10.6439 |
| MLH3 | 5.9981 | 6.6149 | 6.5566 | 7.2083 | 7.2505 | 7.587 |
| TRAPPC3 | 11.2423 | 11.0746 | 11.3781 | 11.9883 | 12.0047 | 12.2092 |
| HSPB7 | 3.7582 | 4.0874 | 4.0351 | 5.2505 | 5.1031 | 4.8834 |
| BZW2 | 10.4895 | 10.5828 | 9.7992 | 11.6616 | 11.2184 | 11.6154 |
| AMDHD2 | 9.1413 | 9.1854 | 11.1211 | 12.2045 | 12.4679 | 13.5131 |
| TMX2 | 9.1716 | 9.437 | 9.7262 | 10.1231 | 10.2283 | 10.4676 |
| TNFRSF12A | 11.133 | 11.3566 | 11.3045 | 13.0377 | 12.7916 | 13.6859 |
| NGRN | 7.5742 | 7.5379 | 9.4218 | 10.4585 | 10.4483 | 10.5525 |
| CYB5R1 | 9.9025 | 9.4133 | 9.7654 | 10.7749 | 10.7986 | 10.4665 |
| SH3TC1 | 6.6799 | 7.5405 | 7.8757 | 8.8429 | 8.7443 | 8.8211 |
| KCTD5 | 5.5286 | 6.2287 | 6.9258 | 8.1035 | 8.4013 | 9.0892 |
| P4HTM | 9.3283 | 9.1027 | 9.3127 | 10.6971 | 10.7446 | 9.8846 |
| SYTL2 | 6.9404 | 7.1202 | 7.0364 | 7.8388 | 8.0144 | 7.617 |
| ZNF654 | 7.2223 | 7.1488 | 7.7198 | 8.5548 | 8.1275 | 8.8731 |
| LSG1 | 10.4346 | 10.6622 | 11.2134 | 11.8286 | 11.9835 | 11.6236 |
| CIDEC | 5.3433 | 5.9069 | 5.4145 | 6.4752 | 7.2543 | 7.7985 |
| NADK | 10.9525 | 11.6038 | 11.2206 | 12.0824 | 12.6528 | 12.5432 |
| NOC4L | 10.0781 | 9.9272 | 10.2064 | 10.6456 | 10.6435 | 10.8953 |
| ZC2HC1C | 6.9655 | 6.9981 | 6.8008 | 7.4816 | 7.6584 | 7.9066 |
| ESRP2 | 8.9021 | 9.6886 | 10.3863 | 10.9381 | 11.6359 | 11.4636 |
| UBTD1 | 8.3399 | 8.502 | 8.2782 | 8.9224 | 9.0672 | 9.2561 |
| ARL14 | 6.3712 | 6.1685 | 6.3187 | 7.0292 | 7.3626 | 7.4506 |
| MAP1LC3B | 10.6529 | 10.4301 | 10.4044 | 11.3258 | 11.0578 | 11.5226 |
| RAB1B | 10.5159 | 10.8491 | 10.5766 | 11.9366 | 11.7869 | 11.9263 |
| UNC119B | 7.3604 | 7.3882 | 7.7578 | 8.4582 | 8.813 | 8.9429 |
| G6PC3 | 9.4793 | 9.851 | 10.1073 | 10.5252 | 11.1011 | 10.9703 |
| FAM171A1 | 5.7998 | 6.342 | 6.699 | 7.4749 | 7.8763 | 7.7254 |
| SFT2D2 | 6.4216 | 6.6865 | 6.6488 | 7.3022 | 7.1989 | 7.0802 |
| AGRN | 10.8111 | 10.4804 | 10.2302 | 11.6817 | 11.4659 | 11.8471 |
TARDBP transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [IL27] and [TARDBP]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
IL27 |
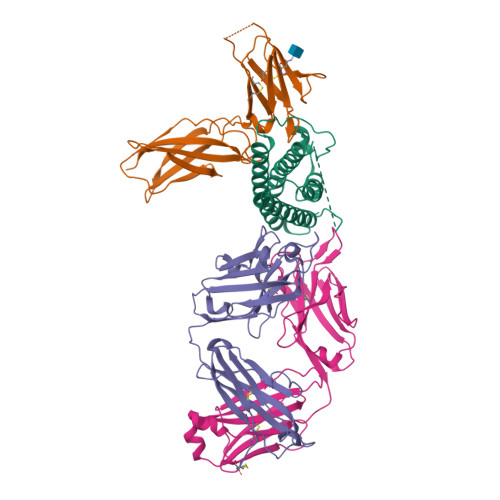
Interleukin-27 subunit alpha
|
243
|
ARTICLE: IL27
[1] Integrative Characterization of the Role of IL27 In Melanoma Using Bioinformatics Analysis.
[3] IL-27 signalling promotes adipocyte thermogenesis and energy expenditure.
[4] The immunobiology of interleukin-27.
[6] The Immunobiology of the Interleukin-12 Family: Room for Discovery.
[7] IL27 T4730C Polymorphism and Serology in Multiple Sclerosis: A Pilot Study.
[8] Tuberculous pleural effusion: diagnosis & management.
[9] A Second-Generation Nanoluc-IL27 Fusion Cytokine for Targeted-Gene-Therapy Applications.
TRANSCRIPTION FACTOR: TARDBP
| Gene | Protein | Amino acids | More |
|
TARDBP |
TAR DNA-binding protein 43
|
414
|