NETWORK ID: CN48945 |
STAG2 transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | PD-0325901 |
| condition dose | 0.12 uM |
| condition time | 24 h |
| CELL LINE | MCF10A |
| CONDITION DETALIS | ||
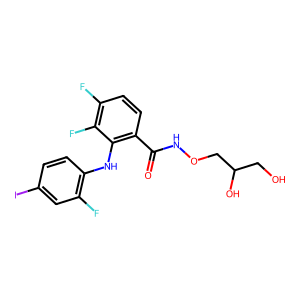 |
ID | BRD-K49865102 |
| name | PD-0325901 | |
| type | trt_cp; compound | |
| molecular formula | C16H14F3IN2O4 | |
| molecular weight | 482.19 g/mol | |
| smiles | ||
| clinical phase | Phase 2 | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| ADCY9 | 7.0247 | 6.7295 | 7.2579 | 8.6527 | 9.2428 | 8.8515 |
| CFB | 6.93 | 9.0408 | 6.784 | 10.4785 | 11.4878 | 10.7239 |
| CDH1 | 6.4965 | 7.2027 | 7.0153 | 9.8274 | 9.1099 | 8.939 |
| CDH3 | 9.5423 | 9.461 | 9.6226 | 10.6718 | 11.1238 | 11.2404 |
| CDKN1A | 10.8528 | 10.8274 | 10.7474 | 11.9068 | 12.0894 | 11.7028 |
| CHRNA3 | 3.786 | 4.1378 | 4.1874 | 5.5321 | 5.6791 | 5.8598 |
| CLDN7 | 9.7397 | 10.18 | 10.1433 | 12.1134 | 11.7891 | 11.8335 |
| CUX1 | 5.1013 | 4.7961 | 5.0861 | 6.8969 | 6.7779 | 6.2517 |
| DHCR24 | 10.6174 | 10.6807 | 10.0738 | 11.7504 | 11.8431 | 11.6512 |
| EPHX1 | 8.7265 | 8.4102 | 8.7268 | 11.6646 | 10.5091 | 10.9901 |
| FABP3 | 6.4952 | 6.9536 | 6.5185 | 8.5924 | 8.7341 | 8.4108 |
| FCER1G | 3.9047 | 3.7718 | 3.6883 | 5.2288 | 4.8024 | 4.6595 |
| FOXO3 | 7.7364 | 7.1391 | 7.6241 | 8.6032 | 8.6576 | 8.5997 |
| GALNT2 | 8.5772 | 7.7598 | 7.8837 | 10.2084 | 9.7747 | 9.527 |
| GLUL | 5.4115 | 6.6172 | 5.4482 | 9.6241 | 8.0871 | 8.385 |
| SFN | 10.7257 | 9.6323 | 10.4683 | 12.2616 | 12.2396 | 11.8564 |
| GSTM3 | 2.124 | 2.2836 | 1.1306 | 3.9267 | 3.7698 | 4.3225 |
| HLA-F | 10.1042 | 10.4806 | 10.381 | 11.9678 | 11.2791 | 11.7449 |
| HLA-G | 8.5233 | 8.7587 | 8.7812 | 9.5165 | 9.7236 | 9.5838 |
| HSPB1 | 10.5626 | 10.4364 | 10.2384 | 13.9998 | 14.0633 | 14.3089 |
| IGF2R | 9.2074 | 9.3244 | 9.2766 | 10.73 | 10.6544 | 10.7004 |
| IL6R | 4.2073 | 4.6423 | 4.3225 | 5.6017 | 5.3176 | 5.3905 |
| IRF6 | 8.8257 | 9.1429 | 9.1175 | 10.6645 | 10.5624 | 11.3939 |
| JUP | 10.8453 | 10.3616 | 10.5908 | 12.9734 | 11.7911 | 12.7912 |
| KRT13 | 4.8946 | 4.6024 | 5.297 | 7.1785 | 6.4317 | 7.5751 |
| KRT16 | 5.8421 | 4.8228 | 6.0087 | 8.5367 | 7.3777 | 7.5733 |
| KRT17 | 9.1264 | 8.3123 | 9.4847 | 13.3001 | 11.3712 | 12.1715 |
| TACSTD2 | 11.867 | 12.1227 | 11.453 | 13.7378 | 14.2149 | 13.2854 |
| MAF | 7.2772 | 7.083 | 7.2353 | 8.1958 | 8.3872 | 8.9721 |
| MUC1 | 6.7814 | 6.5856 | 6.1837 | 8.1854 | 10.1372 | 10.111 |
| MYCL | 4.002 | 4.1548 | 4.42 | 5.7689 | 5.1002 | 5.8147 |
| PIK3C2B | 6.0429 | 6.2433 | 6.6438 | 8.6959 | 8.7312 | 8.3565 |
| POR | 5.1486 | 4.9636 | 5.3048 | 6.9915 | 6.4837 | 6.3288 |
| PPL | 9.3018 | 9.1173 | 9.4596 | 12.4292 | 11.9744 | 12.4497 |
| RGS2 | 10.0098 | 9.9769 | 9.9057 | 13.5866 | 12.5246 | 12.1612 |
| S100A9 | 11.6374 | 11.6253 | 11.4919 | 13.6153 | 14.9208 | 14.0106 |
| SPINT1 | 6.7134 | 7.1204 | 7.7834 | 9.4113 | 10.1731 | 9.9261 |
| SRP14 | 11.2777 | 11.4665 | 11.4743 | 12.5877 | 12.1542 | 12.6317 |
| SULT1A2 | 7.4675 | 7.2488 | 7.3884 | 8.5396 | 8.5207 | 8.6078 |
| STX4 | 9.472 | 9.4847 | 9.231 | 11.8659 | 10.9832 | 11.2525 |
| SULT1A1 | 7.4391 | 7.2874 | 7.3657 | 9.0535 | 9.1869 | 9.1073 |
| TAP1 | 8.0285 | 8.4349 | 8.3379 | 9.5771 | 9.1771 | 9.7742 |
| THBS3 | 7.6154 | 8.3671 | 7.3549 | 9.7528 | 9.4769 | 9.082 |
| TPM1 | 8.3664 | 8.5758 | 8.798 | 10.2005 | 10.9269 | 10.3468 |
| TUFT1 | 7.1645 | 7.6949 | 7.3383 | 9.5923 | 8.6673 | 9.4352 |
| UBE2H | 6.699 | 6.13 | 6.966 | 7.7254 | 8.466 | 8.4714 |
| CREG1 | 12.7547 | 12.5827 | 12.8888 | 14.5351 | 13.8146 | 14.0668 |
| TAX1BP1 | 9.3092 | 9.1737 | 9.2072 | 10.2996 | 10.277 | 10.1091 |
| MPZL1 | 10.4394 | 10.5664 | 10.3222 | 11.473 | 11.7751 | 11.8583 |
| STX8 | 10.0165 | 9.2827 | 9.8472 | 10.8212 | 11.0185 | 11.0122 |
| LITAF | 10.9034 | 11.1998 | 10.2598 | 13.0054 | 12.4209 | 12.402 |
| NPEPPS | 9.7804 | 9.9993 | 9.8148 | 10.9226 | 11.043 | 11.2791 |
| ISG15 | 4.8758 | 4.8694 | 4.8319 | 6.5069 | 7.2416 | 7.4766 |
| PTPRU | 6.9738 | 7.4685 | 7.3773 | 8.7499 | 9.2002 | 8.6432 |
| PDZK1IP1 | 4.8175 | 4.9316 | 5.1796 | 8.5668 | 9.6772 | 8.0731 |
| VAT1 | 8.121 | 7.4461 | 8.3554 | 9.8745 | 10.0093 | 9.8565 |
| SMPDL3A | 8.3144 | 8.5382 | 8.3974 | 9.7144 | 10.4016 | 10.4578 |
| DUSP10 | 7.1987 | 6.6855 | 6.5146 | 8.2566 | 8.3773 | 7.9869 |
| OPN3 | 9.4016 | 9.0195 | 9.1318 | 11.4456 | 11.5289 | 11.3052 |
| MXRA5 | 2.989 | 3.5654 | 2.1093 | 4.7863 | 6.041 | 5.9883 |
| FBXO2 | 5.9602 | 6.0443 | 5.7768 | 7.6085 | 7.5759 | 6.869 |
| SLCO3A1 | 7.0316 | 7.8478 | 7.0468 | 9.4526 | 9.2357 | 9.4124 |
| PYCARD | 9.237 | 9.1284 | 8.8862 | 12.4538 | 11.9558 | 12.1809 |
| FAM8A1 | 7.2916 | 7.2569 | 7.5507 | 8.4073 | 8.8204 | 8.8096 |
| CYB5R1 | 8.2653 | 8.0503 | 8.7077 | 10.4035 | 9.6573 | 10.5286 |
| SYT17 | 8.4921 | 8.3522 | 7.8069 | 9.7058 | 9.781 | 9.9166 |
| WNT4 | 4.3085 | 4.3859 | 4.3265 | 5.3479 | 5.358 | 5.197 |
| HCFC1R1 | 9.324 | 10.1883 | 10.0358 | 11.9088 | 11.2608 | 11.7298 |
| SUSD4 | 5.4506 | 5.6187 | 5.5774 | 7.0055 | 6.5548 | 6.4042 |
| BSDC1 | 9.5139 | 9.179 | 9.4093 | 10.4972 | 10.5209 | 10.4921 |
| LMBRD1 | 7.422 | 7.2015 | 7.3374 | 8.1508 | 8.8281 | 8.5206 |
| PAK6 | 5.7082 | 5.1711 | 5.5502 | 6.6593 | 6.604 | 6.5586 |
| RAB25 | 6.2843 | 5.5438 | 5.8501 | 7.7042 | 7.9726 | 7.5765 |
| S100A14 | 10.8271 | 10.5213 | 11.6816 | 13.2376 | 13.0956 | 13.6627 |
| WFDC1 | 4.49 | 5.7202 | 5.3613 | 7.2018 | 7.0862 | 7.0776 |
| PERP | 9.2342 | 8.9188 | 10.0885 | 11.6178 | 11.0263 | 12.176 |
| MMP28 | 6.9783 | 7.302 | 7.0775 | 8.4849 | 7.9738 | 8.1549 |
| PGBD5 | 5.9831 | 5.7864 | 6.038 | 7.1082 | 7.5876 | 7.1319 |
| ZC3H12A | 7.1975 | 7.5525 | 7.0884 | 8.8715 | 8.6209 | 8.3057 |
| AHNAK2 | 9.4986 | 9.5942 | 10.3055 | 12.1788 | 14.9208 | 12.9361 |
STAG2 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [PD-0325901] and [STAG2]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
MAP2K1 |
Dual specificity mitogen-activated protein kinase kinase 1
|
393
|
ARTICLE: PD-0325901
TRANSCRIPTION FACTOR: STAG2
| Gene | Protein | Amino acids | More |
|
STAG2 |
Cohesin subunit SA-2
|
1231
|