NETWORK ID: CN6161 |
SREBF1 transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | SB-590885 |
| condition dose | 3.33 uM |
| condition time | 24 h |
| CELL LINE | HEPG2 |
| CONDITION DETALIS | ||
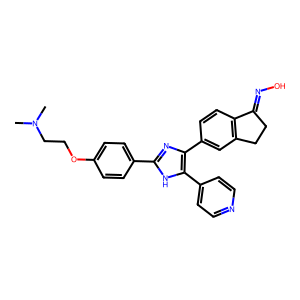 |
ID | BRD-K24496482 |
| name | SB-590885 | |
| type | trt_cp; compound | |
| molecular formula | C27H27N5O2 | |
| molecular weight | 453.5 g/mol | |
| smiles | ||
| clinical phase | None | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| TSR1 | 11.0264 | 10.9658 | 10.9215 | 10.1654 | 9.604 | 9.4467 |
| REXO2 | 10.5285 | 9.9478 | 9.7643 | 8.2827 | 9.5011 | 8.3317 |
| PFDN2 | 11.4649 | 11.5894 | 11.7377 | 10.3751 | 10.7252 | 10.948 |
| TNFRSF12A | 10.3047 | 10.4306 | 9.6489 | 8.9634 | 9.0629 | 7.7466 |
| RBM7 | 8.7201 | 8.2687 | 8.5089 | 7.6626 | 7.589 | 7.6984 |
| CDCA4 | 8.0077 | 8.6788 | 8.0343 | 6.9287 | 7.105 | 6.3294 |
| LYRM4 | 11.6389 | 10.4279 | 10.3459 | 10.1909 | 8.6597 | 8.3651 |
| HEATR1 | 10.3769 | 9.6518 | 10.2855 | 8.9298 | 9.3522 | 8.2218 |
| FNDC3B | 10.4539 | 9.9473 | 9.6913 | 8.9551 | 8.9563 | 8.9107 |
| PUS1 | 8.9699 | 9.11 | 9.0632 | 8.0373 | 7.7486 | 8.3367 |
| SLC35F2 | 8.4212 | 7.6901 | 7.5093 | 6.0696 | 6.3393 | 6.4644 |
| RPL26L1 | 9.9457 | 9.4442 | 8.7483 | 8.4713 | 7.9349 | 7.6314 |
| PAK1IP1 | 10.3975 | 10.6001 | 10.8249 | 8.9613 | 8.8585 | 9.4005 |
| ZNHIT6 | 9.3315 | 9.2342 | 8.8548 | 8.4558 | 7.4579 | 7.447 |
| FASTKD1 | 8.6549 | 8.6373 | 9.1506 | 8.0654 | 8.0035 | 7.5514 |
| RRP15 | 11.2943 | 10.0732 | 10.4655 | 9.2154 | 9.0799 | 9.3374 |
| SLC17A9 | 7.7372 | 8.1139 | 7.7777 | 7.2233 | 6.6199 | 5.9795 |
| ZNF587 | 7.4743 | 7.8992 | 8.6166 | 7.0115 | 6.2685 | 6.9481 |
| P3H1 | 9.1628 | 9.7016 | 9.3529 | 8.3385 | 8.8719 | 8.1117 |
| MYL12B | 11.7019 | 11.7256 | 11.4194 | 10.182 | 10.8239 | 10.4995 |
| DENR | 10.6635 | 10.4408 | 10.1216 | 8.5385 | 9.124 | 8.6879 |
| GLS | 11.6042 | 11.9265 | 9.7861 | 9.575 | 8.7336 | 8.5704 |
| CRISPLD2 | 6.503 | 5.328 | 4.0732 | 3.2606 | 3.662 | 3.6917 |
| METTL5 | 11.199 | 10.9376 | 11.3207 | 10.6119 | 9.7034 | 9.7669 |
| CORO1C | 10.3503 | 10.516 | 9.3795 | 8.2194 | 9.1105 | 8.3477 |
| SEH1L | 11.9231 | 10.888 | 11.0933 | 9.8415 | 9.0829 | 8.2423 |
| SPATA5L1 | 8.6774 | 8.4898 | 9.444 | 7.805 | 7.6172 | 7.851 |
| RDH14 | 8.6946 | 8.6579 | 8.0909 | 7.783 | 6.6715 | 7.4822 |
| KCNE4 | 5.4071 | 5.0598 | 5.9054 | 4.0745 | 4.2461 | 3.9178 |
| EIF4G2 | 12.7225 | 12.1336 | 11.7196 | 10.7696 | 11.2299 | 10.7977 |
| ZNF146 | 9.1924 | 9.1503 | 8.9022 | 8.1692 | 8.1855 | 8.3821 |
| HSP90B1 | 12.3263 | 12.3758 | 12.42 | 10.9602 | 11.1765 | 11.228 |
| DSP | 9.2165 | 9.2919 | 8.3082 | 7.1423 | 8.2202 | 7.0269 |
| CSRP1 | 11.1288 | 11.5163 | 11.6731 | 10.5773 | 10.4079 | 9.5784 |
| LDHA | 13.7814 | 12.7573 | 13.5943 | 11.784 | 12.4854 | 11.3079 |
| PHB | 13.4414 | 13.726 | 12.6606 | 11.0438 | 12.6486 | 10.6289 |
| PPP1CC | 12.3692 | 12.5693 | 12.2269 | 11.5075 | 11.4158 | 11.6051 |
| RAN | 14.0588 | 13.7096 | 14.4158 | 13.2766 | 12.507 | 12.76 |
| ANXA5 | 13.8439 | 13.5329 | 12.5532 | 12.7467 | 11.2484 | 10.3685 |
| ODC1 | 10.4108 | 11.2295 | 10.8995 | 8.3216 | 8.2494 | 7.8409 |
| PPP1CA | 10.8547 | 9.9564 | 9.6998 | 8.7378 | 9.4601 | 8.1874 |
| NOP56 | 14.962 | 14.5805 | 14.0439 | 13.3331 | 13.492 | 12.7836 |
| HSBP1 | 10.2788 | 9.7238 | 10.7533 | 9.506 | 8.8447 | 8.6062 |
| ALDOA | 9.2468 | 9.2989 | 8.4234 | 6.6285 | 8.2406 | 6.2786 |
| ARF4 | 9.747 | 9.4058 | 8.6688 | 8.4371 | 8.1645 | 7.8498 |
| THBS1 | 8.6489 | 8.7144 | 9.2688 | 7.2034 | 6.8832 | 8.0492 |
| PSMA7 | 13.5835 | 13.5038 | 13.4006 | 12.2856 | 13.0111 | 12.1618 |
| SSB | 11.9714 | 12.2348 | 12.2606 | 11.1426 | 11.4993 | 10.9907 |
| ITPR3 | 7.879 | 7.2808 | 7.5138 | 6.8617 | 6.2578 | 5.718 |
| ENO1 | 11.835 | 11.0381 | 11.8184 | 9.9399 | 10.7129 | 9.1444 |
| PSMD13 | 10.9833 | 11.1529 | 10.9255 | 9.3258 | 10.5636 | 9.4417 |
| HNRNPAB | 12.6027 | 12.9622 | 12.5463 | 11.7594 | 11.7552 | 11.3166 |
| EBNA1BP2 | 10.8008 | 10.8003 | 10.5584 | 9.2166 | 9.0313 | 8.9918 |
| DHX15 | 11.6406 | 11.7202 | 10.282 | 9.8552 | 9.5208 | 8.6574 |
| NAP1L4 | 11.5333 | 11.4069 | 11.5216 | 9.9655 | 10.9393 | 10.3497 |
| TALDO1 | 10.3819 | 10.2419 | 9.9641 | 8.8476 | 9.7337 | 8.3407 |
| SRM | 12.0085 | 11.2974 | 11.7328 | 10.0224 | 10.6549 | 9.6746 |
| NME1 | 14.7565 | 14.2448 | 15 | 13.1342 | 13.3609 | 11.7898 |
| FAT1 | 10.6303 | 10.2999 | 10.4419 | 9.3962 | 9.2203 | 8.6603 |
| IFITM1 | 5.5791 | 5.2558 | 4.8952 | 3.7411 | 3.3243 | 3.533 |
| IER3 | 12.1682 | 12.0323 | 11.6895 | 8.8088 | 8.8178 | 8.0716 |
| PNP | 11.229 | 11.489 | 11.5813 | 9.9505 | 10.5054 | 9.6913 |
| MTHFD2 | 11.4596 | 11.159 | 11.1923 | 10.4517 | 10.2233 | 9.2213 |
| TOMM34 | 10.5773 | 10.3198 | 9.9767 | 8.6991 | 9.5273 | 8.7566 |
| CCT2 | 12.5878 | 12.8221 | 12.6848 | 11.7883 | 11.9381 | 11.1769 |
| IER2 | 10.0651 | 9.8975 | 10.3891 | 8.6278 | 9.4312 | 8.8396 |
| TMED5 | 12.2467 | 11.8534 | 12.1093 | 11.3558 | 10.7317 | 11.3349 |
| CBFB | 10.7693 | 10.1922 | 10.1628 | 9.5011 | 9.3351 | 7.9282 |
| NUTF2 | 11.6983 | 10.0942 | 11.3768 | 9.9605 | 9.6829 | 9.1175 |
| UBE2S | 11.1159 | 10.3989 | 11.2626 | 8.8073 | 10.3963 | 8.1959 |
| INPP1 | 8.7751 | 8.755 | 7.9135 | 7.4615 | 7.0237 | 6.2345 |
| CDC20 | 13.3392 | 13.2984 | 13.6604 | 12.2607 | 12.9534 | 12.2089 |
| FHL2 | 9.178 | 8.6288 | 8.7566 | 8.2481 | 7.4377 | 6.9061 |
| CYBA | 9.0418 | 7.1443 | 7.1107 | 5.4633 | 6.9011 | 5.438 |
| SNRPE | 13.3559 | 12.6965 | 12.8081 | 12.1634 | 11.6424 | 12.0256 |
| ITGB1BP1 | 9.8079 | 9.6174 | 9.7046 | 3.9162 | 4.5499 | 9.0834 |
| MTF2 | 9.7609 | 9.6811 | 9.4403 | 8.8224 | 8.7754 | 8.7168 |
| MYCBP | 9.6515 | 9.2746 | 9.0585 | 8.0518 | 8.6254 | 7.4352 |
| PSMG1 | 11.7349 | 11.0777 | 10.5083 | 10.3054 | 9.1636 | 8.3612 |
| MMD | 9.1274 | 8.6064 | 8.6234 | 7.4435 | 7.3088 | 7.9508 |
| MRPL19 | 12.4022 | 12.2755 | 12.6937 | 11.7109 | 11.2426 | 11.2498 |
| SLC7A6 | 7.0987 | 6.865 | 6.3598 | 5.6552 | 5.6885 | 5.6495 |
| PNO1 | 10.1128 | 10.2402 | 10.5449 | 8.1001 | 7.9929 | 8.9302 |
| E2F3 | 9.0825 | 8.4209 | 8.4768 | 8.0684 | 6.6764 | 7.1582 |
| MRPL12 | 13.7426 | 13.9873 | 14.3362 | 12.7044 | 13.5655 | 12.6325 |
| SLC5A6 | 11.155 | 11.0966 | 10.952 | 10.0767 | 10.5143 | 9.5099 |
| MRPS12 | 13.3944 | 13.8728 | 13.6248 | 12.2417 | 12.7924 | 12.772 |
| FOSL1 | 8.3881 | 8.974 | 8.3859 | 6.6974 | 7.5667 | 7.8138 |
| DPH2 | 8.797 | 8.3879 | 8.6047 | 7.338 | 7.7996 | 7.1703 |
| UCHL3 | 12.097 | 11.1343 | 11.4522 | 9.9391 | 10.3802 | 9.5869 |
| EEF1E1 | 13.1673 | 12.6716 | 13.4529 | 11.3363 | 10.3685 | 11.2775 |
| HSPE1 | 14.8933 | 14.8125 | 14.6263 | 14.0171 | 13.3727 | 13.2519 |
| SNRPG | 12.4044 | 11.2917 | 11.5139 | 10.9584 | 10.2969 | 10.3187 |
| COMP | 3.3303 | 3.2745 | 2.665 | 1.7338 | 1.5666 | 1.642 |
| GAS2 | 9.2696 | 8.5874 | 8.8221 | 7.8585 | 7.9674 | 7.42 |
| NOLC1 | 10.1918 | 10.0145 | 9.3582 | 8.9514 | 8.4621 | 8.4147 |
| MATN3 | 8.4011 | 8.5287 | 8.6838 | 7.3567 | 7.567 | 7.919 |
| CNBP | 11.2515 | 11.677 | 11.4128 | 10.9301 | 10.2951 | 10.3131 |
| PRMT1 | 11.4058 | 11.2442 | 10.553 | 9.1422 | 10.4079 | 8.4385 |
| GLRX | 12.4245 | 12.1313 | 12.6639 | 10.978 | 10.7597 | 10.3214 |
| CPNE1 | 12.4576 | 12.4198 | 12.3701 | 10.9928 | 11.9835 | 11.1092 |
| HSPH1 | 11.8588 | 12.1182 | 11.5406 | 10.7602 | 10.4821 | 10.4212 |
| TFRC | 11.8982 | 12.7086 | 13.3992 | 11.3919 | 10.6765 | 10.4924 |
| PCSK6 | 8.4627 | 8.4919 | 9.1374 | 7.8403 | 7.4202 | 7.6743 |
| MAP2K3 | 8.6906 | 8.366 | 8.2295 | 7.352 | 7.35 | 7.0548 |
| PDIA6 | 14.0697 | 14.2389 | 15 | 12.5109 | 12.7875 | 13.5883 |
| ACOT7 | 9.8018 | 10.3753 | 10.7848 | 8.6848 | 8.8962 | 8.8668 |
| CLIC1 | 13.3576 | 13.1027 | 13.9188 | 11.9598 | 12.6743 | 12.1603 |
| PRDX1 | 12.3562 | 11.9018 | 10.2664 | 9.7949 | 10.1798 | 8.268 |
| YWHAB | 12.4111 | 12.4833 | 12.2827 | 10.5259 | 11.9499 | 11.14 |
| ATIC | 11.7809 | 11.3611 | 12.4804 | 10.4791 | 10.5122 | 10.0908 |
| MRPL3 | 10.8451 | 10.8383 | 11.3595 | 9.7078 | 10.1739 | 9.6057 |
| ATP1B3 | 10.7587 | 9.5475 | 10.4125 | 8.8296 | 9.4542 | 8.2443 |
| DUSP6 | 10.4689 | 10.4018 | 10.7429 | 7.3674 | 6.5399 | 7.4877 |
| DDX18 | 10.979 | 10.6427 | 10.6768 | 9.9532 | 9.6349 | 9.2924 |
| C1QBP | 14.5403 | 14.7224 | 15 | 13.4299 | 13.359 | 13.0977 |
| EIF3J | 9.9004 | 9.7401 | 9.7699 | 8.5346 | 8.0959 | 8.6442 |
| TSPAN4 | 10.8639 | 11.0152 | 10.6516 | 9.3296 | 9.981 | 7.9653 |
| ADCY3 | 5.1915 | 4.8463 | 4.4802 | 3.0505 | 4.0447 | 3.7908 |
| UAP1 | 9.2498 | 8.5646 | 7.5883 | 6.823 | 7.2054 | 5.8613 |
| BANF1 | 10.8193 | 11.0663 | 11.3888 | 10.1104 | 10.3171 | 9.4636 |
| TPM1 | 10.7143 | 10.776 | 9.6527 | 9.1012 | 9.3992 | 8.0412 |
| POLR2L | 13.7236 | 13.4986 | 12.4083 | 10.9698 | 12.4713 | 11.0878 |
| PSMD14 | 11.6239 | 12.0671 | 11.9758 | 10.738 | 9.8988 | 10.5064 |
| IMP4 | 11.9069 | 12.7315 | 13.4696 | 11.0482 | 11.2916 | 11.638 |
| GRPEL1 | 10.6017 | 10.8027 | 9.8017 | 9.2941 | 9.6269 | 8.9795 |
| PPP1R14B | 9.5275 | 9.3084 | 9.5435 | 8.753 | 8.346 | 8.3897 |
| RPIA | 11.9016 | 11.5667 | 11.8024 | 10.7331 | 10.9603 | 10.1369 |
| TFPI | 12.9563 | 11.6503 | 10.2129 | 10.6191 | 8.8401 | 8.2221 |
| PFAS | 11.2962 | 10.5619 | 10.6075 | 10.0001 | 9.1177 | 9.195 |
| RPP40 | 9.9602 | 9.3079 | 8.7982 | 8.3829 | 8.0669 | 7.5498 |
| COL2A1 | 6.5024 | 7.2241 | 6.558 | 5.3329 | 5.127 | 5.2755 |
| F2RL1 | 8.007 | 8.6766 | 8.495 | 6.2747 | 6.5063 | 6.9812 |
| HOMER1 | 7.6666 | 6.1766 | 6.0927 | 5.0222 | 5.475 | 4.7357 |
| DHRS2 | 12.0058 | 12.3783 | 11.4245 | 10.5508 | 10.7153 | 9.0119 |
| BCAT1 | 7.4568 | 6.4009 | 6.1127 | 5.6236 | 4.5341 | 3.9098 |
| WDR43 | 10.6496 | 10.1979 | 9.8833 | 9.2596 | 8.7115 | 8.2084 |
| TM4SF1 | 9.5373 | 10.8687 | 9.5592 | 8.0498 | 7.0244 | 4.6455 |
| GTF3A | 11.1941 | 10.616 | 11.1821 | 10.2748 | 9.9348 | 9.3252 |
| ACP1 | 11.1276 | 11.2013 | 11.6423 | 10.2301 | 10.1875 | 10.5511 |
| FASTKD2 | 10.7807 | 10.8668 | 10.9487 | 10.0968 | 10.0029 | 9.9696 |
| POMP | 12.3922 | 11.7851 | 11.6258 | 10.7415 | 10.5806 | 10.5204 |
| PRMT5 | 11.752 | 11.734 | 11.9376 | 10.494 | 10.5168 | 10.1712 |
| PPA1 | 11.4009 | 11.7048 | 11.2352 | 10.3788 | 10.2863 | 10.2614 |
| CMPK1 | 10.2565 | 9.8829 | 9.1063 | 9.0254 | 8.1533 | 8.0246 |
| AKIRIN1 | 10.1898 | 8.8813 | 8.8369 | 8.0289 | 8.0949 | 7.983 |
| HSD17B11 | 10.7963 | 10.4079 | 10.5281 | 9.724 | 9.0898 | 9.4733 |
| PHLDA1 | 8.7641 | 7.9928 | 7.0296 | 3.5472 | 5.3317 | 5.6223 |
| NDC1 | 9.5248 | 9.7682 | 9.5268 | 8.7858 | 8.3301 | 8.1128 |
| SRPRB | 10.1735 | 9.5072 | 8.3686 | 8.2204 | 7.9042 | 6.9184 |
SREBF1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [SB-590885] and [SREBF1]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
BRAF |
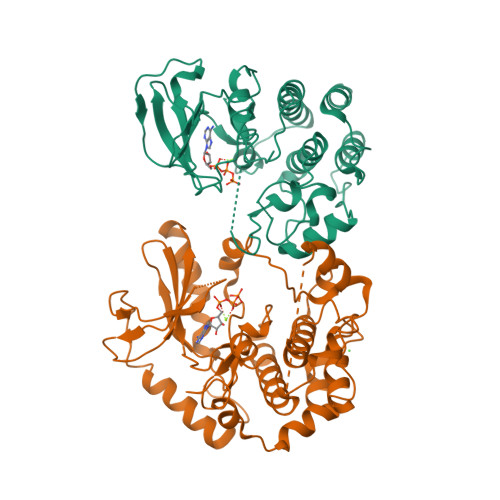
Serine/threonine-protein kinase B-raf
|
766
|
ARTICLE: SB-590885
[3] The identification of potent and selective imidazole-based inhibitors of B-Raf kinase.
TRANSCRIPTION FACTOR: SREBF1
| Gene | Protein | Amino acids | More |
|
SREBF1 |
Sterol regulatory element-binding protein 1
|
1147
|
ARTICLE: SREBF1
[3] Srebf1 Controls Midbrain Dopaminergic Neurogenesis.
[6] Role of AMP-activated protein kinase in mechanism of metformin action.
[7] SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver.
[8] SREBF1 Activity Is Regulated by an AR/mTOR Nuclear Axis in Prostate Cancer.