NETWORK ID: CN70465 |
GREB1 transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | chlorpropamide |
| condition dose | 2.22 uM |
| condition time | 24 h |
| CELL LINE | MCF7 |
| CONDITION DETALIS | ||
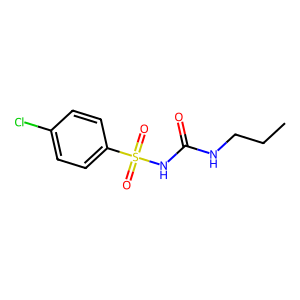 |
ID | BRD-K97746869 |
| name | chlorpropamide | |
| type | trt_cp; compound | |
| molecular formula | C10H13ClN2O3S | |
| molecular weight | 276.74 g/mol | |
| smiles | ||
| clinical phase | FDA Approved | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition |
|---|---|---|---|---|---|
| ABCA3 | 7.1428 | 7.4835 | 7.266 | 8.4181 | 7.8898 |
| ACTA1 | 7.2687 | 7.3644 | 7.4281 | 9.0414 | 8.9021 |
| APBA2 | 7.1435 | 6.8907 | 6.8934 | 7.7696 | 7.5612 |
| ATF3 | 6.3898 | 6.6675 | 6.0349 | 7.484 | 7.0943 |
| ATP2A1 | 6.8211 | 6.1066 | 6.321 | 7.835 | 7.6926 |
| CALML3 | 4.9936 | 5.0469 | 5.0002 | 7.4812 | 5.8053 |
| CASQ1 | 5.7213 | 5.3199 | 5.588 | 6.6521 | 6.2144 |
| CRYAB | 6.2602 | 5.248 | 6.1597 | 7.8898 | 9.0217 |
| ENO3 | 7.0367 | 6.5392 | 6.5378 | 7.6791 | 8.5383 |
| EPHX1 | 8.308 | 7.964 | 7.4412 | 8.7161 | 10.3711 |
| ETFA | 12.2983 | 12.6632 | 12.2692 | 13.4632 | 13.0042 |
| FGF7 | 3.5365 | 3.8245 | 3.503 | 4.2239 | 4.467 |
| ITPKB | 6.8675 | 6.3608 | 6.3255 | 7.5471 | 8.6619 |
| KRT15 | 8.6573 | 9.1475 | 9.0472 | 10.5176 | 10.4239 |
| MMP7 | 7.348 | 6.6087 | 7.001 | 9.1114 | 8.9096 |
| MYBPC1 | 7.9283 | 7.4845 | 8.6113 | 10.4239 | 9.5487 |
| MYH2 | 6.2773 | 5.7327 | 5.9595 | 8.3626 | 7.6086 |
| MYL2 | 6.6027 | 6.0145 | 6.5654 | 8.9324 | 8.2864 |
| NDUFAB1 | 11.3003 | 11.3538 | 11.5124 | 12.0282 | 12.0214 |
| NFYC | 7.4577 | 7.6698 | 7.9396 | 8.2723 | 9.0103 |
| NMT1 | 6.7597 | 6.7996 | 6.7559 | 7.2562 | 7.8294 |
| PDK2 | 7.6077 | 7.7121 | 7.5632 | 8.8146 | 9.0406 |
| PKD1 | 6.9901 | 6.8525 | 6.954 | 7.8799 | 7.9603 |
| PKP1 | 4.9776 | 4.7844 | 4.1272 | 6.6702 | 5.4867 |
| PLIN1 | 5.4802 | 5.7684 | 6.0091 | 7.1585 | 6.7946 |
| PRRX1 | 2.9014 | 3.1649 | 2.7747 | 4.6285 | 4.3476 |
| PPL | 9.1354 | 8.5306 | 7.7232 | 9.5501 | 10.5266 |
| PPP1R1A | 5.0165 | 4.5917 | 3.5936 | 6.3441 | 6.4134 |
| PYGM | 7.0832 | 6.9066 | 7.2101 | 8.3903 | 8.4424 |
| RABGGTA | 7.3378 | 7.4689 | 6.9775 | 8.3494 | 8.194 |
| RDH5 | 6.8809 | 6.976 | 7.4405 | 7.8303 | 8.131 |
| RGS13 | 3.9109 | 3.2316 | 3.3427 | 4.9916 | 4.1826 |
| S100A3 | 5.6329 | 5.5617 | 5.4208 | 6.354 | 6.3973 |
| S100A8 | 9.9481 | 7.5921 | 6.5882 | 12.1886 | 11.7461 |
| S100A9 | 7.3717 | 7.6309 | 7.1573 | 11.5734 | 9.527 |
| SALL2 | 7.795 | 7.5862 | 7.6167 | 8.4226 | 8.5226 |
| CXCL12 | 6.9123 | 8.7145 | 7.019 | 11.0148 | 9.0617 |
| SELL | 3.3263 | 4.0285 | 3.7675 | 5.4966 | 4.478 |
| SP2 | 6.9525 | 7.293 | 7.4459 | 8.1319 | 8.4505 |
| TLR5 | 4.4071 | 4.1993 | 4.9128 | 5.8347 | 5.2392 |
| TNNI1 | 7.3984 | 6.3874 | 6.932 | 8.2438 | 8.0255 |
| TNNI2 | 7.3166 | 6.9113 | 7.8127 | 8.6735 | 8.3254 |
| TPM3 | 7.6769 | 6.6284 | 7.5042 | 8.6351 | 8.787 |
| TULP3 | 9.6769 | 9.1266 | 9.3871 | 10.6699 | 10.6164 |
| CSRP3 | 6.7349 | 6.4158 | 6.1321 | 8.1085 | 7.3592 |
| TCAP | 7.2687 | 7.2138 | 7.2052 | 9.4283 | 8.4661 |
| KMO | 3.9162 | 3.2867 | 2.8481 | 5.6026 | 5.246 |
| LDB1 | 6.6425 | 6.6736 | 6.5766 | 7.228 | 7.2633 |
| AP1G2 | 8.1806 | 8.4759 | 8.0846 | 9.6846 | 8.975 |
| ENDOU | 4.164 | 4.4485 | 4.313 | 5.1031 | 5.2064 |
| SLC7A7 | 4.8261 | 5.2713 | 5.3463 | 6.4865 | 5.7587 |
| RPS6KA5 | 4.7421 | 4.4004 | 4.9891 | 5.8674 | 6.1156 |
| RAPGEF3 | 5.7158 | 5.6993 | 5.22 | 6.2138 | 6.584 |
| DEAF1 | 6.2203 | 6.4247 | 6.113 | 6.9388 | 6.9393 |
| TXNIP | 0 | 2.5239 | 2.7515 | 4.3605 | 4.3347 |
| LYVE1 | 4.9461 | 4.5523 | 4.7806 | 5.6658 | 5.3745 |
| PRR4 | 5.5982 | 5.7776 | 6.2295 | 6.6199 | 6.8581 |
| RAP1GAP2 | 7.2342 | 7.6375 | 7.7871 | 8.3682 | 8.6462 |
| MGRN1 | 8.5428 | 8.7763 | 8.9904 | 9.4546 | 9.7497 |
| PHLDA3 | 7.7692 | 7.2633 | 7.3807 | 8.573 | 8.5204 |
| METTL7A | 5.2021 | 4.0916 | 5.8162 | 8.4269 | 6.8857 |
| DECR2 | 7.5436 | 7.7575 | 7.8587 | 8.4054 | 8.4469 |
| RNF115 | 9.6469 | 9.8079 | 9.8373 | 10.5932 | 10.5037 |
| MINK1 | 6.3785 | 7.6941 | 6.5065 | 8.1479 | 8.6477 |
| UTP18 | 10.9961 | 11.0626 | 10.7263 | 11.6093 | 12.2903 |
| NAGPA | 7.1274 | 7.6156 | 7.6617 | 8.2129 | 8.282 |
| TUBD1 | 9.5698 | 10.1875 | 9.4557 | 10.8009 | 10.785 |
| CHST15 | 4.7731 | 4.726 | 4.6887 | 5.4706 | 5.2622 |
| UFC1 | 8.7994 | 9.8612 | 9.4172 | 10.2788 | 11.1133 |
| GPRC5B | 5.6027 | 5.7478 | 6.606 | 7.8515 | 8.0153 |
| MXRA8 | 4.743 | 5.1872 | 4.2612 | 6.5478 | 5.5058 |
| MANSC1 | 9.9255 | 8.6929 | 10.2164 | 11.0748 | 11.0575 |
| ZNF692 | 8.0131 | 7.8122 | 7.6272 | 8.7794 | 8.4913 |
| HHAT | 5.4045 | 5.2857 | 4.9703 | 5.8471 | 6.0319 |
| METTL3 | 6.22 | 6.1139 | 6.0079 | 6.9392 | 6.9195 |
| PPM1H | 8.315 | 7.5734 | 8.8715 | 9.5121 | 9.4479 |
| MYOZ1 | 6.3747 | 5.9668 | 5.9726 | 7.3427 | 7.1907 |
| FTO | 5.8398 | 6.148 | 6.11 | 6.6169 | 7.3073 |
| ACSS3 | 6.1125 | 6.2062 | 6.411 | 7.153 | 6.8815 |
| NAA16 | 8.754 | 7.8746 | 8.4422 | 9.2812 | 9.3826 |
| MSANTD2 | 5.8214 | 5.4675 | 5.3255 | 6.7322 | 6.4467 |
| PALB2 | 6.7068 | 6.6772 | 6.5911 | 7.7663 | 7.6561 |
| ECHDC3 | 6.2075 | 5.9976 | 6.0873 | 6.9096 | 7.948 |
| CLMN | 4.6099 | 4.7135 | 4.2287 | 5.2447 | 5.5915 |
| MMRN2 | 4.5744 | 4.2371 | 4.6817 | 5.2776 | 5.2095 |
| SYNPO2L | 5.8313 | 5.7369 | 5.7541 | 6.6862 | 6.2719 |
| CDK5RAP3 | 9.234 | 9.4653 | 9.0397 | 10.155 | 10.6219 |
| HYI | 7.4358 | 7.9453 | 7.4999 | 8.3254 | 8.5456 |
| LONP2 | 6.2127 | 6.6043 | 6.3323 | 7.4604 | 7.2834 |
| KIF18B | 7.1511 | 6.9209 | 6.7923 | 7.5217 | 7.8549 |
| ZFC3H1 | 7.09 | 7.3286 | 6.9783 | 8.1052 | 7.6895 |
| FLCN | 5.2294 | 5.2128 | 5.2153 | 5.9693 | 5.8444 |
| FAM174B | 7.8422 | 7.7717 | 7.7245 | 9.1448 | 8.832 |
GREB1 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [chlorpropamide] and [GREB1]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
ABCC8 |
ATP-binding cassette sub-family C member 8
|
1581
|
TRANSCRIPTION FACTOR: GREB1
| Gene | Protein | Amino acids | More |
|
GREB1 |
Protein GREB1
|
1949
|
ARTICLE: GREB1
[1] Uterine mesenchymal tumours: recent advances.
[3] GREB1 regulates PI3K/Akt signaling to control hormone-sensitive breast cancer proliferation.
[4] GREB1 isoform 4 is specifically transcribed by MITF and required for melanoma proliferation.
[5] Role for Growth Regulation by Estrogen in Breast Cancer 1 (GREB1) in Hormone-Dependent Cancers.
[6] GREB1 functions as a growth promoter and is modulated by IL6/STAT3 in breast cancer.
[7] greb1 regulates convergent extension movement and pituitary development in zebrafish.
[10] GREB1 genetic variants are associated with bone mineral density in Caucasians.