NETWORK ID: CN8127 |
E2F4 transcriptional regulatory sub-network
| EXPERIMENTS | |
| condition | MG-132 |
| condition dose | 20 uM |
| condition time | 24 h |
| CELL LINE | MCF7 |
| CONDITION DETALIS | ||
 |
ID | BRD-K60230970 |
| name | MG-132 | |
| type | trt_cp; compound | |
| molecular formula | C26H41N3O5 | |
| molecular weight | 475.6 g/mol | |
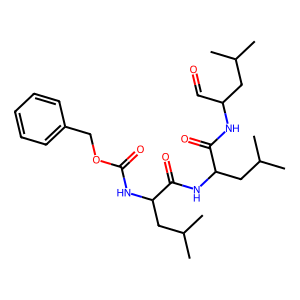
| smiles | ||
| clinical phase | None | |
Differential expressed genes in this transcriptional regulatory sub-network
If you want to view heatmap, you can click [expand]
You can download all DEGs data.
| Symbol | control | control | control | condition | condition | condition |
|---|---|---|---|---|---|---|
| OLFML3 | 6.8692 | 6.7864 | 7.1007 | 5.6544 | 5.3933 | 5.1305 |
| UQCR10 | 14.2084 | 13.815 | 13.2442 | 12.2625 | 10.7776 | 10.7084 |
| MLPH | 13.4784 | 13.461 | 13.5259 | 10.1205 | 8.3674 | 9.2414 |
| ZWILCH | 8.0855 | 8.6374 | 8.2654 | 6.4063 | 5.7138 | 6.0089 |
| GMNN | 12.4243 | 12.4661 | 12.3116 | 10.9366 | 10.8727 | 11.3255 |
| KIF4A | 7.9859 | 8.0058 | 8.0933 | 5.486 | 5.9236 | 5.5981 |
| DARS2 | 9.0735 | 9.5657 | 9.4668 | 7.9935 | 8.1421 | 7.8598 |
| CARHSP1 | 8.8344 | 8.3295 | 9.1744 | 7.448 | 7.013 | 6.9734 |
| DTL | 6.705 | 6.349 | 7.01 | 4.2206 | 4.5457 | 3.8621 |
| SUV39H1 | 7.8735 | 7.4734 | 7.5892 | 6.0126 | 5.8985 | 5.7012 |
| HJURP | 8.0698 | 8.1343 | 8.0912 | 5.9058 | 5.851 | 6.8594 |
| KIF20A | 9.6155 | 9.3163 | 9.2855 | 5.3879 | 5.9349 | 5.2233 |
| FBXO5 | 7.7099 | 7.8429 | 8.0667 | 5.5216 | 5.5178 | 5.6128 |
| CENPU | 9.6406 | 8.688 | 9.3549 | 6.6667 | 6.0355 | 6.0588 |
| RMI1 | 7.6737 | 7.493 | 7.8494 | 5.5348 | 5.6567 | 4.6485 |
| DSCC1 | 7.6091 | 7.3857 | 7.5466 | 6.4843 | 5.8086 | 6.0353 |
| LAGE3 | 8.5658 | 8.0511 | 8.3392 | 5.5091 | 5.9586 | 5.2691 |
| AMDHD2 | 8.2104 | 7.9467 | 7.8706 | 6.2159 | 6.4836 | 6.3363 |
| PBK | 9.0129 | 8.7899 | 9.2987 | 5.696 | 5.4427 | 6.5191 |
| C3orf14 | 15 | 14.5681 | 15 | 10.7216 | 11.6472 | 11.7012 |
| KIF15 | 5.9827 | 7.2394 | 6.9135 | 4.7578 | 4.5985 | 4.8255 |
| ATRAID | 10.3811 | 10.5358 | 9.8343 | 8.9938 | 8.9131 | 8.5938 |
| GRHL2 | 7.1622 | 7.074 | 7.4538 | 5.6314 | 5.8085 | 6.0203 |
| SHCBP1 | 7.6599 | 8.2575 | 7.9712 | 6.39 | 5.8933 | 6.7806 |
| RAD54B | 8.3905 | 8.5422 | 8.5963 | 7.2226 | 6.9482 | 6.9625 |
| NEIL3 | 6.8249 | 7.4652 | 7.3732 | 5.8137 | 5.863 | 5.6301 |
| NCAPG2 | 9.007 | 8.9851 | 8.9071 | 7.0545 | 6.9174 | 7.1857 |
| THAP10 | 9.5437 | 9.2658 | 9.137 | 8.0142 | 7.2561 | 8.0194 |
| ECT2 | 9.7115 | 10.5221 | 8.9892 | 7.1628 | 6.4509 | 7.3067 |
| ASPM | 8.3801 | 7.7548 | 7.8064 | 4.0887 | 4.9111 | 5.1996 |
| ZDHHC4 | 7.8606 | 7.9529 | 7.7407 | 5.8337 | 6.5407 | 6.1904 |
| DEPDC1 | 7.3067 | 7.5628 | 7.7009 | 5.5843 | 6.0196 | 6.0385 |
| C1orf112 | 7.5392 | 7.6251 | 7.3031 | 6.0462 | 5.3064 | 6.0091 |
| KIF18A | 6.6825 | 7.4051 | 7.3316 | 5.4406 | 5.2921 | 5.6608 |
| CDCA3 | 8.6381 | 9.6065 | 9.1152 | 6.7786 | 6.4439 | 7.3432 |
| BRIP1 | 7.6796 | 7.4087 | 7.7805 | 6.6177 | 5.6722 | 5.7065 |
| KIAA1324 | 8.2571 | 9.0491 | 8.1636 | 6.9289 | 7.175 | 6.8006 |
| TRMT5 | 12.487 | 12.4006 | 12.2342 | 9.712 | 9.6796 | 7.867 |
| MCM4 | 8.8728 | 8.76 | 8.9699 | 7.6951 | 6.8742 | 7.2024 |
| KIF18B | 7.4074 | 6.9569 | 7.0678 | 5.2512 | 5.5132 | 4.9851 |
| RACGAP1 | 10.1482 | 9.2148 | 8.9307 | 6.3127 | 5.8559 | 6.8097 |
| CELSR2 | 7.5158 | 7.3016 | 7.3611 | 6.1381 | 6.137 | 5.7893 |
| HCFC1R1 | 10.4882 | 9.8747 | 10.2701 | 8.7864 | 8.8516 | 8.0327 |
| XBP1 | 11.425 | 11.0709 | 11.236 | 7.7715 | 7.4934 | 8.0937 |
| STMN1 | 8.6129 | 8.1417 | 9.0371 | 7.1457 | 6.8574 | 7.0331 |
| TOP2A | 11.4747 | 12.1139 | 11.5537 | 6.0802 | 7.608 | 7.7526 |
| SH3BGRL | 10.5769 | 10.7291 | 11.0961 | 7.8928 | 8.9279 | 8.0965 |
| HNRNPF | 9.183 | 9.0864 | 9.0622 | 8.1933 | 7.4954 | 7.4933 |
| MCM3 | 9.75 | 9.2672 | 9.5883 | 7.4817 | 7.9878 | 8.1738 |
| SMC4 | 9.1461 | 8.9118 | 9.4239 | 7.1517 | 7.3217 | 8.0257 |
| DLG5 | 7.8375 | 8.2019 | 7.6269 | 6.4865 | 6.1744 | 6.2652 |
| MYBL2 | 9.2446 | 8.9308 | 8.3539 | 5.7714 | 6.2069 | 6.104 |
| TWF1 | 9.6601 | 10.1194 | 9.5228 | 8.2599 | 8.2391 | 7.7758 |
| TP53 | 10.2861 | 9.7103 | 9.4183 | 8.3543 | 7.5431 | 7.3988 |
| NCAPD2 | 6.8664 | 7.0932 | 6.7073 | 5.2963 | 5.3702 | 5.6898 |
| NET1 | 8.282 | 7.7144 | 8.9544 | 6.5805 | 6.2215 | 6.4421 |
| CDC25B | 11.0812 | 11.4367 | 11.2213 | 9.069 | 9.0634 | 9.25 |
| RRM2 | 8.9529 | 9.1356 | 8.478 | 6.3124 | 6.7031 | 7.4707 |
| IMPDH2 | 8.4861 | 8.7002 | 8.7699 | 6.8238 | 6.5314 | 7.1576 |
| PSRC1 | 9.4446 | 9.4732 | 9.7881 | 7.3464 | 6.7942 | 6.855 |
| CKS1B | 11.7056 | 11.5322 | 10.6429 | 9.3688 | 9.3897 | 9.3071 |
| BIRC5 | 11.7074 | 11.7882 | 11.5537 | 6.7934 | 7.731 | 6.5019 |
| ARHGAP1 | 10.1993 | 9.8792 | 9.8385 | 8.5203 | 8.8582 | 8.861 |
| KIF22 | 10.3006 | 10.5208 | 10.3196 | 8.7926 | 9.1172 | 8.9656 |
| PLK1 | 9.2193 | 9.2047 | 9.5376 | 6.2942 | 5.2715 | 5.6472 |
| TK1 | 9.6416 | 9.1875 | 9.8612 | 7.1344 | 8.06 | 7.3884 |
| SERPINA3 | 3.2823 | 4.6486 | 4.479 | 0.9286 | 0 | 0 |
| ERBB3 | 8.6713 | 9.1241 | 9.0849 | 6.5169 | 6.2215 | 6.2802 |
| PRSS23 | 10.8537 | 11.0909 | 10.9866 | 9.3746 | 9.4901 | 9.5929 |
| GALE | 9.49 | 9.1938 | 8.9728 | 7.532 | 6.9359 | 7.2612 |
| FOXM1 | 11.0321 | 10.1034 | 10.4312 | 7.5895 | 7.0861 | 7.1057 |
| S100A13 | 12.6231 | 12.5635 | 12.4633 | 11.1901 | 10.9409 | 10.969 |
| CCNB2 | 11.557 | 11.8536 | 11.4897 | 9.2143 | 8.9743 | 9.1407 |
| LIG1 | 9.028 | 8.7051 | 8.7947 | 6.2453 | 6.3123 | 6.0775 |
| UBE2C | 11.6382 | 10.6795 | 11.205 | 7.1711 | 7.4934 | 6.6236 |
| RNASEH2A | 9.8446 | 9.8186 | 9.5043 | 7.8499 | 7.2173 | 7.469 |
| TIMELESS | 9.5604 | 9.5382 | 8.9092 | 6.1983 | 6.3883 | 6.3936 |
| CDK1 | 10.1616 | 9.5622 | 10.2309 | 5.0974 | 5.5245 | 5.4982 |
| DTYMK | 9.4867 | 9.8175 | 9.4682 | 7.939 | 8.3326 | 7.4603 |
| SLC35A1 | 10.0202 | 10.2162 | 9.8783 | 8.891 | 8.6601 | 8.4952 |
| MAD2L1 | 10.1238 | 10.1561 | 10.0226 | 6.1914 | 6.6521 | 7.6655 |
| DDB2 | 9.4359 | 9.3985 | 9.3117 | 7.5519 | 8.077 | 7.7833 |
| ALG8 | 8.2554 | 8.5963 | 8.6305 | 7.3225 | 7.2584 | 7.246 |
| PTTG1 | 10.3696 | 9.1661 | 9.3881 | 6.9614 | 7.1738 | 6.6787 |
| GPRC5B | 5.7467 | 6.0938 | 6.3449 | 4.4609 | 4.0673 | 2.7206 |
| DLGAP5 | 9.7376 | 10.6221 | 9.6362 | 6.535 | 6.8018 | 6.3109 |
| VRK1 | 8.2429 | 8.5026 | 7.9387 | 6.6626 | 6.5932 | 7.0169 |
| CLDN3 | 10.8963 | 10.5458 | 10.1717 | 8.383 | 7.7922 | 8.7361 |
| TSNAX | 9.5924 | 9.9691 | 9.6099 | 8.3072 | 8.1384 | 8.5187 |
| ZWINT | 11.7524 | 10.4922 | 11.732 | 8.6774 | 9.6292 | 8.1422 |
| AURKA | 7.1141 | 6.8363 | 7.3631 | 5.0974 | 5.3496 | 5.4558 |
| RAD51AP1 | 8.8613 | 8.7871 | 9.1542 | 6.6898 | 7.0833 | 7.0215 |
| WASF1 | 7.4518 | 7.5238 | 7.7325 | 6.2786 | 5.1578 | 6.1118 |
| SMC2 | 7.607 | 6.9416 | 7.3186 | 5.7591 | 5.5834 | 6.2018 |
| GTSE1 | 8.5034 | 7.8728 | 7.9951 | 5.2897 | 5.37 | 5.3199 |
| KIF11 | 7.7058 | 7.9077 | 7.5945 | 5.2938 | 5.5472 | 5.9381 |
| BRCA1 | 5.9833 | 5.8522 | 5.4826 | 4.637 | 4.4587 | 3.9169 |
| NEK2 | 8.8061 | 8.5247 | 8.14 | 4.9616 | 4.5748 | 4.7548 |
| TROAP | 7.9707 | 7.3138 | 7.6865 | 6.092 | 5.8593 | 5.6424 |
| KIF23 | 7.2712 | 8.146 | 7.0591 | 5.4975 | 5.1549 | 5.9674 |
| MYB | 6.6752 | 7.237 | 7.1011 | 2.417 | 4.8449 | 3.6285 |
| TTK | 8.0713 | 7.4693 | 7.2699 | 5.745 | 5.6658 | 5.1237 |
| TFF1 | 13.2863 | 15 | 13.0234 | 8.3798 | 9.0358 | 7.9047 |
| LSM6 | 7.0193 | 6.8434 | 6.6853 | 5.2729 | 5.2037 | 5.5362 |
| ITGB3BP | 9.2335 | 9.307 | 9.2367 | 8.024 | 8.1943 | 7.6473 |
| DNALI1 | 8.5687 | 8.2191 | 8.5234 | 7.0902 | 6.7682 | 7.4086 |
| PLS1 | 8.9308 | 8.845 | 8.5834 | 6.7237 | 7.1434 | 7.3279 |
| ESR1 | 11.1448 | 10.7487 | 10.1094 | 8.1591 | 6.625 | 4.8067 |
| TRMT2B | 7.4048 | 7.7949 | 7.0089 | 6.238 | 5.8483 | 5.925 |
| CHEK1 | 7.8735 | 7.8148 | 7.9847 | 6.9207 | 6.3769 | 6.3363 |
| SCYL3 | 6.3951 | 6.1005 | 6.2832 | 4.693 | 4.3571 | 4.7504 |
| KIF14 | 7.4061 | 7.2474 | 7.2309 | 5.0201 | 5.1411 | 5.0715 |
| HOXC6 | 5.0277 | 3.953 | 3.9092 | 1.6619 | 1.7505 | 1.9174 |
| AKT1 | 9.9371 | 9.9118 | 9.9269 | 8.8424 | 8.6776 | 8.7833 |
| HMMR | 10.2394 | 9.9122 | 9.86 | 6.7652 | 7.3441 | 6.0556 |
| CENPF | 6.8748 | 7.1732 | 7.2107 | 4.3593 | 5.0244 | 3.9831 |
| HMGN2 | 12.1796 | 11.7777 | 11.6706 | 9.9846 | 9.643 | 8.8287 |
| HMGB2 | 9.2915 | 10.2115 | 10.5802 | 7.4152 | 6.797 | 7.0874 |
| POLE3 | 9.4671 | 9.1343 | 9.9709 | 7.8241 | 7.4238 | 7.1191 |
| DUT | 9.6993 | 9.1172 | 9.7423 | 8.0408 | 7.5025 | 7.4936 |
| TAX1BP3 | 11.2895 | 10.3449 | 10.4144 | 8.9883 | 9.1549 | 8.7523 |
| AGR2 | 11.2513 | 9.5784 | 8.9727 | 3.9221 | 5.9106 | 6.3727 |
| AZGP1 | 11.0617 | 12.2117 | 13.1214 | 6.4474 | 8.5077 | 7.4807 |
| PATZ1 | 8.3841 | 7.8836 | 7.8127 | 6.6617 | 6.2007 | 6.8945 |
| AURKB | 8.4533 | 8.4473 | 8.3848 | 4.637 | 5.4165 | 4.9219 |
| TMX1 | 10.1092 | 9.8254 | 9.8677 | 8.7971 | 8.1281 | 8.2793 |
| NSL1 | 7.3666 | 7.2438 | 6.8681 | 6.0698 | 5.7773 | 5.7233 |
| RPA3 | 7.9824 | 7.4734 | 7.6361 | 5.3171 | 5.3345 | 5.4861 |
| BUB1 | 8.911 | 9.4073 | 8.2023 | 5.462 | 5.3853 | 6.1721 |
| CDKN3 | 10.7504 | 9.4795 | 10.1032 | 7.6312 | 7.2726 | 7.7918 |
| TGFB3 | 6.2525 | 6.1589 | 5.8753 | 4.4891 | 4.3175 | 4.6362 |
| SPC25 | 7.1344 | 7.5563 | 7.7779 | 5.2012 | 5.4815 | 6.2627 |
| SLC25A1 | 10.8792 | 10.6339 | 11.0416 | 9.2214 | 8.9648 | 9.6602 |
| TPX2 | 9.7301 | 8.624 | 8.8112 | 6.0316 | 6.204 | 6.379 |
| CHEK2 | 7.9467 | 7.6793 | 7.8978 | 6.6774 | 6.3359 | 6.1627 |
| ANXA9 | 6.7712 | 6.9766 | 6.8989 | 4.4564 | 5.0722 | 4.4798 |
| BDH1 | 8.3669 | 7.8254 | 7.6361 | 5.9864 | 6.176 | 6.1236 |
| LSM5 | 11.1706 | 11.2636 | 11.168 | 6.5133 | 9.1938 | 6.566 |
| KPNA2 | 11.5934 | 12.0286 | 11.2637 | 10.2737 | 9.7847 | 10.1058 |
| ARL6IP1 | 9.3653 | 9.0624 | 9.1883 | 7.4469 | 7.0964 | 6.6505 |
| MKI67 | 6.9083 | 7.2637 | 6.9317 | 4.2497 | 5.6372 | 5.0632 |
| CBX5 | 9.5345 | 8.8839 | 9.7696 | 7.1016 | 7.9412 | 7.3382 |
| WEE1 | 10.6891 | 10.7517 | 10.5534 | 8.6629 | 8.7741 | 8.9635 |
| C2CD5 | 10.1892 | 10.3481 | 9.9166 | 8.5259 | 8.3583 | 8.0018 |
| NCAPH | 8.4439 | 8.2694 | 8.4845 | 6.6912 | 6.5658 | 6.868 |
| FANCI | 7.347 | 6.8576 | 7.0019 | 5.1222 | 5.6984 | 5.7955 |
| ERI2 | 8.6074 | 7.9268 | 9.0742 | 6.279 | 6.547 | 6.6875 |
| OIP5 | 7.079 | 7.8785 | 7.5724 | 5.6117 | 6.3719 | 5.5018 |
| MYBL1 | 7.9673 | 7.0254 | 7.3529 | 4.3942 | 4.6279 | 4.695 |
| AKR7A2 | 11.6626 | 11.2082 | 11.3063 | 8.9527 | 9.0466 | 9.6408 |
| SPDEF | 11.7312 | 11.3972 | 11.0519 | 7.863 | 8.0912 | 6.6236 |
| GGCT | 11.0457 | 10.4527 | 11.6774 | 8.2237 | 9.2642 | 8.2329 |
| ADI1 | 7.8603 | 7.8643 | 7.7106 | 6.3363 | 6.4144 | 6.7865 |
| EVL | 6.8467 | 7.0263 | 7.0704 | 5.4325 | 4.8549 | 5.7123 |
| CYBRD1 | 5.6754 | 5.9796 | 6.0907 | 4.5914 | 4.6932 | 4.3431 |
| IARS2 | 11.0048 | 11.357 | 10.8195 | 9.5224 | 9.5236 | 9.4183 |
| KLHDC2 | 11.4573 | 11.7572 | 11.3813 | 10.3714 | 10.3667 | 10.1388 |
| STUB1 | 13.6538 | 13.3811 | 13.5743 | 12.4243 | 12.3652 | 11.9303 |
| DCXR | 10.2748 | 11.1749 | 10.5231 | 8.4016 | 8.9838 | 8.8133 |
| NUSAP1 | 8.7805 | 8.0511 | 8.4261 | 5.4657 | 6.3087 | 7.0072 |
| ASF1B | 9.3541 | 9.2006 | 9.6048 | 7.2257 | 7.0028 | 6.8585 |
E2F4 transcriptional regulatory sub-network
The possible genes regulated by the transcription factor were obtained by calculation, and the results were displayed in the form of network
More information of [MG-132] and [E2F4]. Click to[expand]
Curated target protein/gene of this condition/drug
Protein target or gene target of this condition curated by previous studies and their associated KEGG pathways are shown below
| Target Gene | Protein | Amino acids | More |
|
PSMB1 |

Proteasome subunit beta type-1
|
241
|
ARTICLE: MG-132
[1] MG-132 attenuates cardiac deterioration of viral myocarditis via AMPK pathway.
[2] MG-132 interferes with iron cellular homeostasis and alters virulence of bovine herpesvirus 1.
[3] MG-132 treatment promotes TRAIL-mediated apoptosis in SEB-1 sebocytes.
[4] Effect of MG-132 on myofibrillogenesis and the ubiquitination of GAPDH in quail myotubes.
[5] MG-132 reduces virus release in Bovine herpesvirus-1 infection.
[7] MG-132 reverses multidrug resistance by activating the JNK signaling pathway in FaDu/T cells.
[8] Effects of proteasome inhibitor MG-132 on the parasite Schistosoma mansoni.
[9] The Role of NF-kB Inhibitors in Cell Response to Radiation.
TRANSCRIPTION FACTOR: E2F4
| Gene | Protein | Amino acids | More |
|
E2F4 |
Transcription factor E2F4
|
413
|
ARTICLE: E2F4
[3] E2f4 is required for intestinal and otolith development in zebrafish.
[4] E2F4 function in G2: maintaining G2-arrest to prevent mitotic entry with damaged DNA.
[5] E2F4 regulates cell cycle to mediate embryonic development in pigs.
[6] Novel functions for the transcription factor E2F4 in development and disease.
[7] E2F4 Promotes the Proliferation of Hepatocellular Carcinoma Cells through Upregulation of CDCA3.